When the coronavirus disease 2019 (COVID-19) first emerged and spread across the globe, many governments were forced to enact costly and restrictive measures to reduce the transmission of the disease. These measures included social distancing guidelines, business, public space closures, and even full lockdowns and stay-at-home orders. As mass vaccination schemes in developed countries began to come to fruition, these measures were mostly dismantled. However, with the rise of dangerous severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) variants of concern (VoCs) such as Delta and Omicron, several European countries have reintroduced them. These VoCs show different traits that make them threatening, including but not limited to evasion of vaccine and infection-induced immunity, higher transmission rates, and stronger ACE2 binding.
In new research, scientists from Adaptive Biotechnologies have been investigating the impact of spike protein mutations on vaccine efficiency for the Omicron variant. The study is currently available on the medRxiv* preprint server while awaiting peer review.
 Study: Immunosequencing and epitope mapping reveal substantial preservation of the T cell immune response to Omicron generated by SARS-CoV-2 vaccines. Image Credit: NIAID
Study: Immunosequencing and epitope mapping reveal substantial preservation of the T cell immune response to Omicron generated by SARS-CoV-2 vaccines. Image Credit: NIAID

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Background
The spike protein is key to the pathogenicity of SARS-CoV-2. It is formed of two subunits that must be cleaved by a host protein – S1 and S2. S1 contains a receptor-binding domain (RBD) that can bind to angiotensin-converting enzyme 2 (ACE2), permitting viral cell entry, while S2 is responsible for mediating membrane fusion. Most variants of concern show mutations in the RBD, which is the target of most vaccines and can lead to changes in vaccine efficacy.
The Study
The researchers mapped the resolution of 199 HLA class I epitopes across the spike protein, aiming for resolution to exact n-mers or short overlapping stretches. They also obtained the resolution of the full set of HLA class II epitopes to around 50 amino acid windows. Combining this with data obtained from immunosequencing the TCR repertoires from over 5000 individuals following SARS-CoV-2 infection or vaccination allowed them to determine the relative fraction of T cells induced by each antigen as well as the HLA association of T cell clones.
This data was used to examine the effect of mutations on the cellular immune response. The researchers assumed that the mutations were not selected to avoid the cellular immune response; confidence in this assumption is supported by the diversity of HLA molecules that present the epitopes.
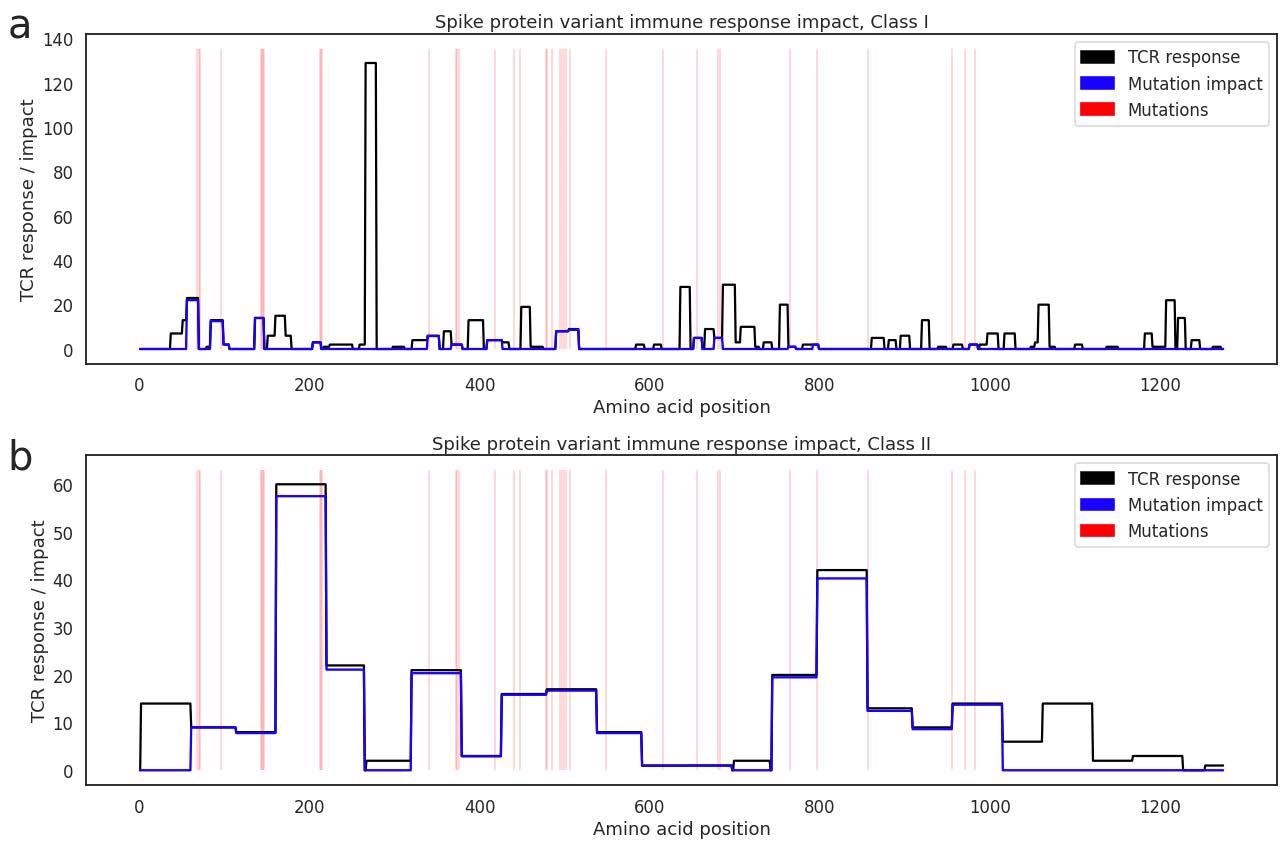
Depicting size of the Class I (a) and Class II (b) T cell response to each epitope range across the spike protein in our antigen map (black line), with positions of Omicron mutations marked in red. The blue line estimates the amount of the cellular immune response that is offtarget within each epitope window when challenged with Omicron.
To test this assumption, they reviewed the data to try and find any enrichment off class I or II HLAs for prior variants or Omicron, looking across all antigen mutations. No statistically significant enrichments from the Delta or Omicron variants were observed within the spike, but the class II HLA association (DQA1*05:05+DQB1*03:01 was found to connect to the nucleocapsid phosphoprotein antigen mutation R203M in Delta, and R203K/G204R in Omicron. This was identified from both repertoire-focused and antigen-focused analyses.
The second assumption the scientists made related to the probability of a mutation in an epitope or the flanking sequences to epitopes preventing the epitope from being presented, or otherwise affecting binding. This would cause vaccine/infection-induced TCRs to show as off-target. They decided that a conservative assumption would allow any coding mutation that spanned a single epitope to negatively affect all TCRs' binding to that single epitope.
Taking these assumptions into account, they determined that mutations in the Omicron variant could affect as much as 21% of the class I-restricted and 33% of the class II-restricted cellular immune response that is elicited by SARS-CoV-2 vaccines. Not all mutations will be impacted by antigen presentation, just as not all mutations will be impacted by recognition by specific TCRs. Still, the authors present the assessment score as an upper bound on the impact these mutations can have. When examining T cell responses that are targeted to non-spike regions, if induced by natural infection, the impact on the Omicron variants is below 5% as the mutations are concentrated in the spike gene and relatively rare in the rest of the genome.
Conclusion
The authors highlight that their study has shown that over two-thirds of the cellular immune response will be preserved, which likely indicates a similar reduction in spike-specific cellular immunity. Luckily, it seems T cell response is less affected, although there is no conclusion on how effective the T cell response is against the disease. It seems likely that T-cell response cannot prevent infection but can fight the disease once established. This information could be key for vaccine manufacturers and healthcare workers and help inform future public health policy.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.