An interesting preprint research paper describes structural changes resulting from the multiple mutations found in the recent Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 is the causative agent behind the coronavirus disease 2019 (COVID-19) pandemic. In addition, the research describes the consequent effects of these changes on Omicron’s infectivity and immune evasion capabilities.
The COVID-19 pandemic has caused millions of deaths and hundreds of millions of infections. The attempts to stop the spread of the virus by national and regional lockdowns have caused severe financial stress and economic hardship, affecting almost every area of daily life. Despite the rollout of vaccines and the development of monoclonal antibodies against the virus, the emergence of new variants with immune escape characteristics presents a formidable challenge to the goal of freeing the world of this plague.
The Omicron variant of concern (VOC) of SARS-CoV-2 not only has the most significant number of mutations seen so far among all the variants but is spreading with unprecedented speed and escapes humoral immunity much more effectively than any other variant so far. This is thought to be due to a large number of spike mutations with this VOC.
The current paper, available on the bioRxiv* preprint server, describes the results of examining the mutated structure of the Omicron antigens using a combination of techniques, including cryo-electron microscopy and X-ray crystallography. In addition, surface plasmon resonance (SPR) studies were used to assess the binding affinity of therapeutic monoclonal antibodies (mAb) in use at present for Omicron RBD. This revealed the reason for the increased infectivity of the Omicron variant, in the presence of electrostatic shifts in the interactions between the spike and the host angiotensin-converting enzyme 2 (ACE2) receptor.
The study also shows how spike-receptor binding, involving the engagement of the receptor-binding domain (RBD) of the viral spike to the host receptor, as well as to the mAbs, is impaired by the change in structure due to the numerous spike mutations. This was done by examining the complexes formed by virus RBD binding to the broadly neutralizing sarbecovirus S309 (the parent mAb of sotrovimab).
The Omicron VOC spike protein has 37 mutations compared to the wildtype virus, compared to the 19 in the Alpha and Delta VOCs, the earlier variants that similarly swept the world. There are 15 and 11 mutations in the Omicron RBD and N-terminal domain (NTD), respectively, linked to marked impairment of neutralization by antibodies elicited by natural infection or prior vaccination when re-exposed to the virus.
The 15 spike RBD mutations of Omicron do not affect ACE2 binding in humans but do confer mouse ACE2 recognition capacity. This antigenic shift, as it is called, also caused most currently available mAbs to lose neutralizing activity against Omicron, with the notable exception of S309 and the cocktail of COV2-2196/COV2-2130 (cilgavimab/tixagevimab parent). While the former lost potency by 2-3-fold, the latter showed 12-200-fold lower potency against the pseudovirus or authentic virus in neutralizing assays.
To further understand this threat to pandemic control, the investigators examined the prefusion stabilized Omicron spike ectodomain trimer in complex with S309 and S2L20, which binds the RBD and NTD respectively. The antibody-binding fragments (Fab) in complex with the RBD and ACE2 were specifically subjected to cryo-EM and X-ray crystallography, respectively.
What Did the Study Show?
The Omicron VOC has many mutations found in earlier variants, both in the RBD and the NTD. The presence of 8 additional mutations outside the RBD, NTD and furin cleavage site of the spike protein makes the Omicron a far more complex subject of study than the earlier VOCs. Four of these eight mutations result in new electrostatic interactions between the core helices of the S2 subunit of the spike and the S1 subunit.
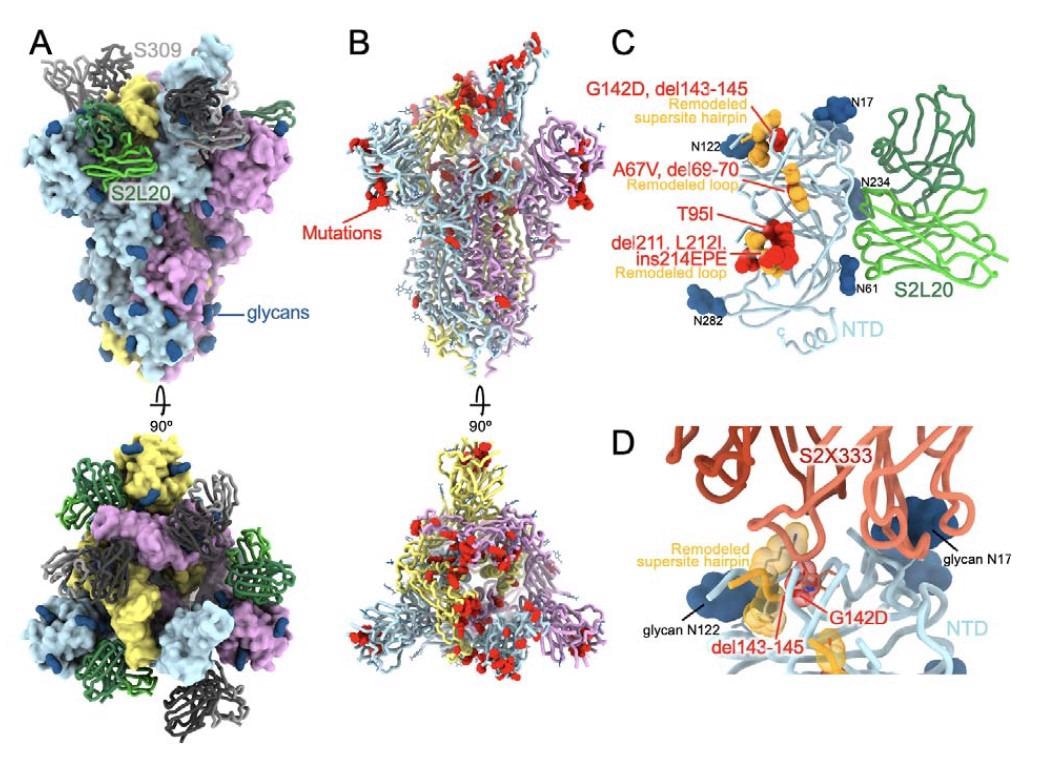
CryoEM structure of the SARS-CoV-2 Omicron S reveals remodeling of the NTD antigenic supersite. (A) Surface rendering in two orthogonal orientations of the Omicron S trimer with one open RBD bound to the S309 (grey) and S2L20 (green) Fabs shown as ribbons. (B) Ribbon diagrams in two orthogonal orientations of the S trimer with one open RBD with residues mutated relative to Wuhan-Hu-1 shown as red spheres (except D614G which is not shown). In panels A-B, the three S protomers are colored light blue, pink or gold. (C) The S2L20-bound Omicron NTD with mutated, deleted or inserted residues rendered or indicated as red spheres. Segments with notable structural changes are shown in orange and labeled. (D) Zoomed-in view of the Omicron NTD antigenic supersite highlighting incompatibility with recognition by the S2X333 mAb (15) (used here as an example of prototypical NTD neutralizing mAb). N-linked glycans are shown as dark blue surfaces.
Another mutation, L981F, enhances the hydrophobic packing of the residues. These mutations occur in regions adjacent to the prefusion stabilizing 2P mutations used in all the currently approved three vaccines available in the USA.
The Omicron mutations may produce more interactions between the two spike subunits and a change in the way the S1/S1 cleavage site is processed in the presence of the N679K and P681H mutations. This might account for the increased effector function of antibodies elicited by natural infection or vaccination, or mAbs with Fc-mediated effector function, by reducing the shedding of the S1 subunit that precedes viral entry into the host cell.
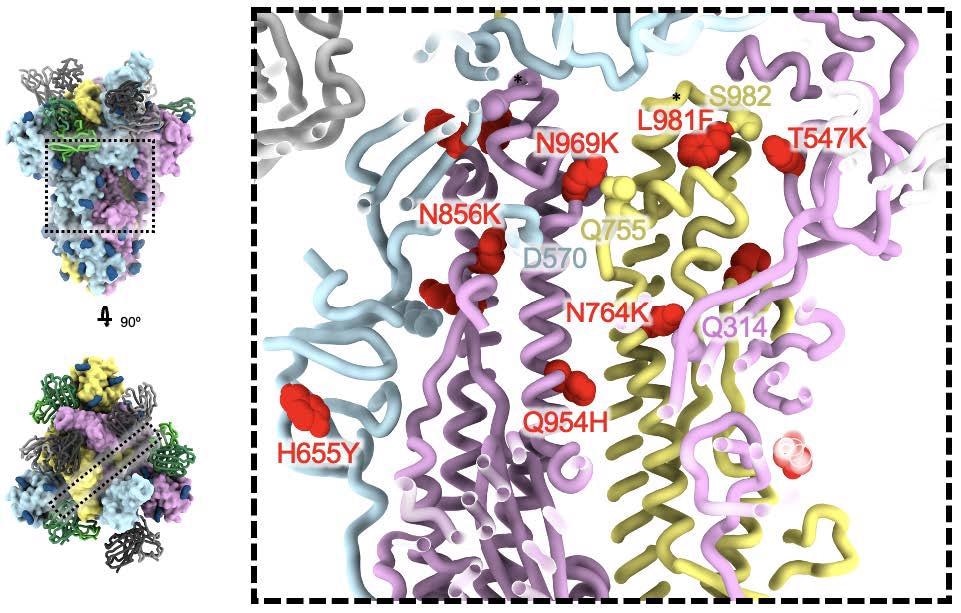
SARS-CoV-2 Omicron S fusion machinery mutations. A cross section through the core of the spike glycoprotein is shown (the location of this slice on the spike glycoprotein is shown on the left). Mutations T547K, H655Y, N764K, N856K, Q954H, N969K, and L981F are shown as red spheres; residues these mutations interact with are shown as spheres colored as the protomer they belong to. Black asterisks show the position of residues involved in the prefusion-stabilizing 2P mutations (K986P and V987P) used in all three vaccines deployed in the US.
The RBD is the immunodominant antigen, with several distinct antigenic sites to which neutralizing antibodies are directed with diverse potencies and breadth of neutralization. The scientists found that electrostatic interactions were lost in the presence of mutations such as K417N, E484A and Q493R, with steric hindrance with REGN10933 being introduced.
Conversely, G446S caused a steric clash with REGN10987, completely inhibiting Omicron RBD binding to this mAb. Several such clashes were observed to dampen antibody-mediated neutralization of the Omicron RBD by COV2-2196 and COV2-2130, compared to the wild-type virus.
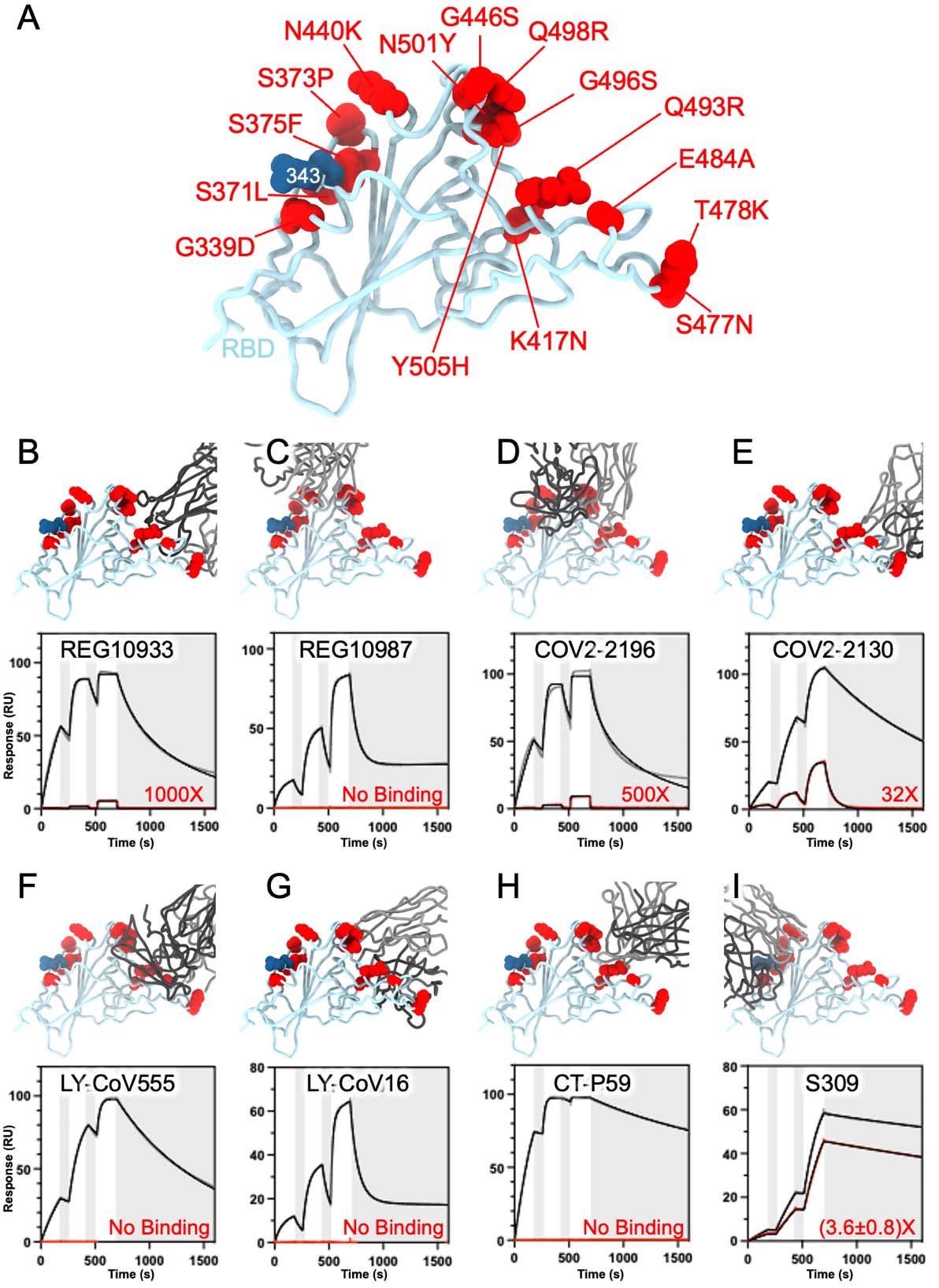
SARS-CoV-2 Omicron RBD mutations promote escape from a panel of clinical mAbs. A, Ribbon diagram of the RBD with residue mutated relative to the Wuhan-Hu-1 RBD shown as red spheres. The N343 glycan is rendered as blue spheres. B-I, Zoomed-in view of the Omicron RBD superimposed to structures of the RBD bound to REGN10933 (B), REGN10987 (C), COV2-2196 (D), COV2-2130 (E), LY-CoV555 (F), LY-CoV16 (G), CT-P59 (H) or S309 (I). Binding of the Wuhan-Hu-1 (gray line) or Omicron (red line) RBD to the corresponding mAb was evaluated using surface plasmon resonance (single-cycle kinetics) and is shown at the bottom. The black line is a fit to a kinetic model. The decrease in affinity between Wuhan-Hu-1 and Omicron binding is indicated in red.
With LY-CoV555, the E484A mutation inhibited hydrogen bonding between the RBD and the heavy and light chains of the mAb, while Q493R prevents binding via steric clashes, again. The heavy chain of LY-CoV16 cannot bind the Omicron RBD because of the loss of multiple electrostatic interactions between these molecules with the introduction of K417N.
The triplet of K417N E484A and Q493R mutations also abolish binding with the CT-P59 mAb by steric hindrance and the loss of electrostatic contacts. Interestingly, the results obtained using these techniques agree with those from deep mutational scanning that predicted the effects of mutations at each of the residues of the RBD.
Why does S309 retain its activity? The Omicron G339D and N440K mutations occur very near or within the S309 epitope on antigenic site IV, but both introduce side chains that cause moderate disruption binding with the mAb, with a corresponding 2-3-fold fall in neutralizing potency of the VOC.
The N501Y mutation found in the Alpha and Beta VOCs did not cause efficient binding of the mouse ACE2 receptor, but this effect is found in the Omicron variant. This could be due to the presence of the Q493R mutation that has electrostatic interactions with the mouse ACE2, and which becomes fixed in serial mouse passages. The result is a mouse-adapted virus SARS-CoV-2 MA10.
What Are the Implications?
“This work defines the molecular basis for the broad evasion of humoral immunity exhibited by SARS-CoV-2 Omicron and underscores the SARS-CoV-2 S mutational plasticity and the importance of targeting conserved epitopes for vaccine and therapeutics and design.”
The loss of neutralizing activity with clinical mAbs and mAb cocktails when confronted with Omicron RBD, except for S309, is a significant challenge to COVID-19 mitigation and treatment. Approximately one in ten isolates of Omicron have the R346K substitution that is linked to evasion of the C135 mAb, in combination with the N440K mutation present in all isolates. However, R346K does not impair S309 binding.
S309 was obtained from a recovered SARS-CoV patient (infected in 2003), but C135 from a recovered SARS-CoV-2 patient. The former thus presented an excellent opportunity to find broadly neutralizing sarbecovirus antibodies that target epitopes that are highly conserved in this family.
The mutational constraints on such sites prevent the ready emergency of immune-evading variants. Meanwhile, the identification of such antibodies offers hope for the development of broadly neutralizing sarbecovirus vaccines.
“These efforts offer hope that the same strategies that contribute to solving the current pandemic will prepare us for future putative sarbecovirus pandemics.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
McCallum, M. et al. (2021). Structural Basis Of SARS-Cov-2 Omicron Immune Evasion and Receptor Engagement. bioRxiv preprint. doi: https://doi.org/10.1101/2021.12.28.474380, https://www.biorxiv.org/content/10.1101/2021.12.28.474380v1
- Peer reviewed and published scientific report.
McCallum, Matthew, Nadine Czudnochowski, Laura E. Rosen, Samantha K. Zepeda, John E. Bowen, Alexandra C. Walls, Kevin Hauser, et al. 2022. “Structural Basis of SARS-CoV-2 Omicron Immune Evasion and Receptor Engagement.” Science 375 (6583): 864–68. https://doi.org/10.1126/science.abn8652. https://www.science.org/doi/10.1126/science.abn8652.