The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by the rapid worldwide outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has claimed more than 5.55 million lives.
Scientists have reported that the spike (S) protein of SARS-CoV-2 contains two domains, namely, S1 and S2, which establish the viral infection. S1 subunit contains the receptor-binding domain (RBD), which binds to the receptor angiotensin-converting enzyme 2 (ACE2) expressed in host cells. The primary function of the S2 subunit is to promote the fusion of viral–host membranes and, thereby, facilitating viral entry into the host cell.
Prior studies have indicated that the S protein is the principal target of cellular and humoral responses elicited by natural infection. To date, all the available COVID-19 vaccines and therapeutics have been developed to target the S protein of SARS-CoV-2.
Since the emergence of SARS-CoV-2 in Wuhan, China, in late December 2019, it has evolved owing to recombination, insertion, and deletion events in the viral genome. This has led to the emergence of many SARS-CoV-2 variants that have been classified as variants of concern (VOC) (e.g., Alpha, Beta, Delta, Omicron), variants of interest (VOI) (e.g., Epsilon, Iota, Kappa), by the World Health Organization (WHO). SARS-CoV-2 variants, classified as VOCs, have been characterized as more virulent and transmissible than the original SARS-CoV-2 strain. Additionally, these variants can evade immune responses elicited by natural infection or vaccination.
Study: Antigenicity of the Mu (B.1.621) and A.2.5 SARS-CoV-2 Spikes. Image Credit: Lightspring/Shutterstock
Mu and A.2.5 Variants
In March 2021, scientists identified a new variant from Colombia, which was named the Mu (B.1.621) variant, and was categorized as a VOI. Mu contains several mutations in the S protein, including 146N, T95I, Y144T, and Y145S mutations in the N-terminal domain (NTD), R346K, E484K N501Y in RBD, and P681H at the S1/S2 interface. A surge in the numbers of SARS-CoV-2 Mu strain was found in Québec (Canada) during the summer, which slowed down the transmission of the Delta strain.
In 2021 the Delta variant became the dominant circulating strain in most countries of the world and caused severe SARS-CoV-2 infection. This occurrence raised the question of the possible evolutionary advantage of this variant.
Researchers also identified another SARS-CoV-2 variant, which probably spread from Central America to the Quebec province in Canada, and named it A.2.5. This variant contained deletions 141–143 at the NTD and the L452R mutations in RBD. This variant belonged to the A.* lineage that was abundantly found during the early phase of the pandemic and had been replaced by the B lineage, subsequently. In early 2021, many countries reported a resurgence of the A.* lineage.
Québec experienced two outbreaks associated with A.2.5 with an increased secondary attack rate. Researchers performed sequencing of A.2.5 and observed a convergent acquisition of the D614G and L452R signature mutations of the Beta and Delta variant, respectively. In addition, they reported a mutational jump of twenty-three mutations, a characteristic feature of the majority of the VOCs.
A New Study
To gain a better understanding of the antigenic properties of Mu and A.2.5 Spikes, Canadian scientists evaluated their capacity to interact with ACE2 and performed binding and neutralization assays with plasma from vaccinated individuals. The research is published in the journal Viruses. The researchers compared the spike glycoproteins of Mu and A.2.5 variants with Beta and Delta Spike variants.
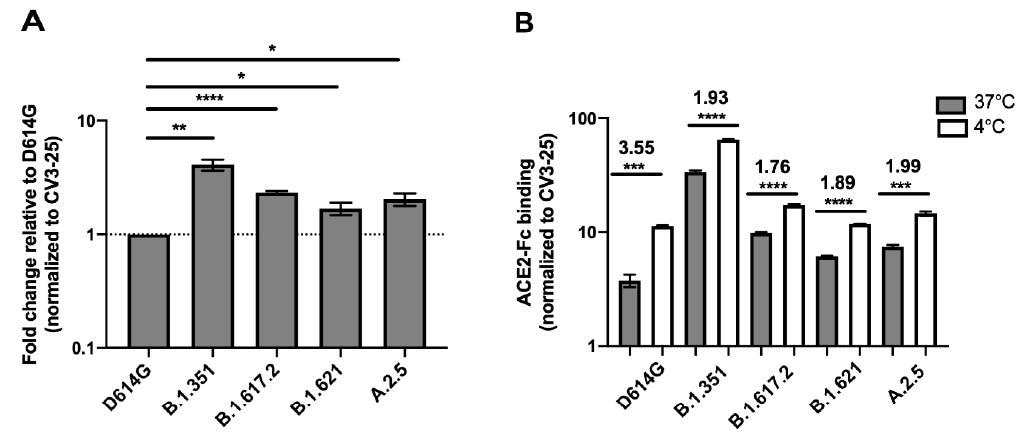
Spike glycoproteins interaction with ACE2. HEK 293T cells were transfected with the indicated SARS-CoV-2 Spike variants. 48 h post-transfection, cells were stained with ACE2-Fc or with CV3-25 Ab and analyzed by flow cytometry. ACE2-Fc binding to the different full Spike variants is presented as a ratio of ACE2 binding of D614G Spike (A). The graph represented ACE2-Fc binding to the different full Spike variants at 37 °C and 4 °C (B). For each Spike variant, statistical significance and fold changes of ACE2 binding at 4 °C vs. 37 °C is indicated in (B). ACE2-Fc binding was normalized to CV3-25 binding in each experiment and at each indicated temperature. Error bars indicate means ± SEM. Statistical significance was performed using Mann–Whitney U test (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001).
The findings of this study are in line with previous studies that revealed a considerable increase in ACE2-Fc binding. Scientists reported that although both Mu and A.2.5 bound ACE2 better than D614G, the binding capacity was lower than the Beta and Delta Spikes. However, it was not clear if the differential binding rate contributed to the transmission rates of these variants.
Previous studies by the same team of researchers showed that temperature variation impacted the RBD–ACE2 interface by modulating the Spike trimer conformation. This study further stated that at cold temperature (4 °C), the binding between S protein of VOCs/VOIs and ACE2 increased significantly. Similarly, scientists found that both Mu and A.2.5 variants revealed better ACE2 interaction at lower temperatures.
Mutation at the RBD region enhanced the affinity of both the variants to the ACE2 receptor at a lower temperature, as it promoted thermodynamic stability of the ACE2–Spike–RBD complex. This is not the case for every mutation. For instance, N501Y mutation is not associated with thermodynamic stability. Previous studies have revealed that N501Y mutation, which is found in Alpha, Beta, and Gamma, can independently impact ACE2 interaction irrespective of temperature.
Importantly, researchers observed that Mu and A.2.5 Spikes were detected in the plasma of vaccinated naïve and previously-infected individuals. The neutralizing capacity of Mu was observed to be lower compared to A.2.5.
Conclusion
The authors indicated that specific mutations and lower temperatures enhance viral transmissibility and replication. This study further demonstrated the significance of measuring critical parameters, such as plasma recognition, ACE2 interaction, etc., of all newly emerging SARS-CoV-2 variants.