The B.1.1.529 variant of SARS-CoV-2, officially named the Omicron variant by the World Health Organization (WHO), was initially reported in November 2021 and was quickly identified as a significant variant of concern. The problem with this specific variant is that it is currently circulating even among doubly vaccinated individuals, with massive public health ramifications.
We already know that SARS-CoV-2 uses its spike glycoprotein for host cell entry by recognizing the angiotensin-converting enzyme 2 (ACE2) receptor, which was the rationale behind vaccine development. However, the Omicron variant spike protein harbors 37 mutations and represents the most mutated variant of this virus to date.
Therefore, putting into context the repercussions of these mutations for ACE2 receptor binding and antibody evasion process is pivotal for the development of efficacious therapeutics intended to limit the spread of the Omicron variant, but also those that may emerge later.
And this task was in the focus of a research group from the Department of Biochemistry and Molecular Biology of the University of British Columbia, as well as from the BC Center for Disease Control Public Health Laboratory in Vancouver, Canada.
From cryogenic electron microscopy to neutralization studies
In their recent paper published in the top-tier journal Science, one of the main methodological approaches was the use of cryogenic electron microscopy (cryo-EM) to appraise the structure of the Omicron spike protein in unprecedented detail.
Furthermore, in order to precisely measure the influence of Omicron spike glycoprotein mutations on binding affinity to the ACE2 receptor on human cells, the researchers have performed surface plasmon resonance studies and subsequently compared the resulting apparent binding affinities to spike glycoproteins of wild-type and Delta strains.
They have also utilized a panel of neutralizing monoclonal antibodies that include antibodies directed to the receptor-binding domain (RBD) and N-terminal domain (NTD) of SARS-CoV-2 spike glycoprotein to appraise the impact of Omicron RBD and NTD mutations on monoclonal antibody escape.
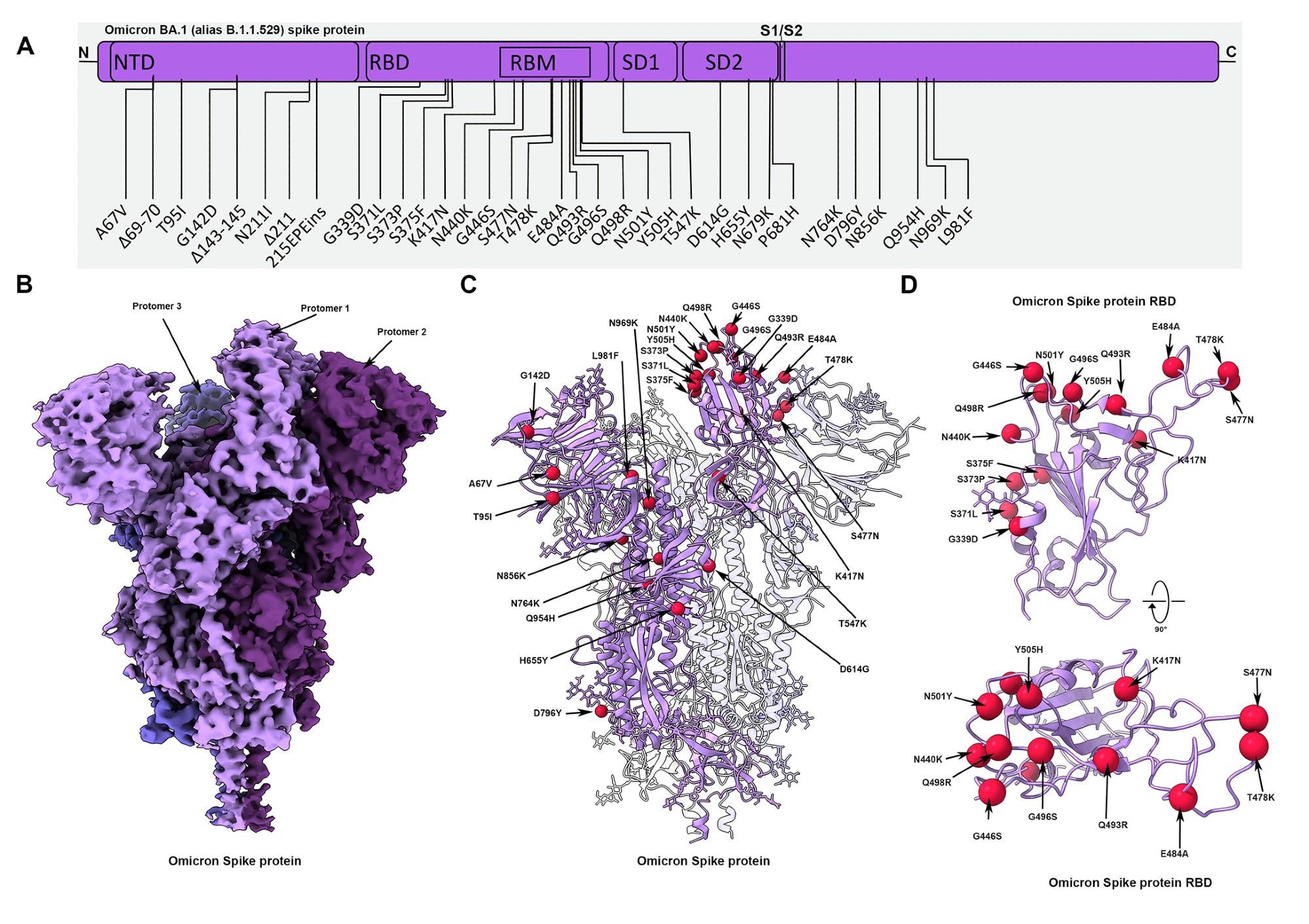 Cryo-EM structure of the Omicron spike protein.(A) A schematic diagram illustrating the domain arrangement of the spike protein. Mutations present in the Omicron variant spike protein are labeled. (B) Cryo-EM map of the Omicron spike protein at 2.79 Å resolution. Protomers are colored in different shades of purple. (C) Cryo-EM structure of Omicron spike protein indicating the locations of modeled mutations on one protomer. (D) The Omicron spike receptor-binding domain (RBD) shown in two orthogonal orientations with Cα positions of the mutated residues shown as red spheres.
Cryo-EM structure of the Omicron spike protein.(A) A schematic diagram illustrating the domain arrangement of the spike protein. Mutations present in the Omicron variant spike protein are labeled. (B) Cryo-EM map of the Omicron spike protein at 2.79 Å resolution. Protomers are colored in different shades of purple. (C) Cryo-EM structure of Omicron spike protein indicating the locations of modeled mutations on one protomer. (D) The Omicron spike receptor-binding domain (RBD) shown in two orthogonal orientations with Cα positions of the mutated residues shown as red spheres.
Mutations that reinstitute binding efficiency
In short, a structural analysis of the Omicron variant spike glycoprotein with cryo-ER in complex with human ACE2 revealed new salt bridges and hydrogen bonds that were formed by mutated residues R493, S496, and R498 in the RBD with ACE2.
This is an important finding, as these interactions seem to compensate for other Omicron mutations which are known to reduce ACE2 binding affinity (such as K417N) – giving rise to similar biochemical ACE2 binding affinities for both Delta and Omicron variants.
In addition, the cryo-EM structure of the complex between spike glycoprotein and the ACE2 receptor exposes a structural explanation for this phenomenon. More specifically, interactions that involve novel mutations in the Omicron variant at residues 493, 496, 498, and 501 appear to reimpose ACE2 binding efficiency that would be lost due to other mutations (like aforementioned K417N).
Therefore, the remarkably pervasive mutations in the Omicron variant spike glycoprotein appear to spark a rather broad antibody escape relative to previously emerged and circulating variants of SARS-CoV-2.
The study has also revealed the reduction in neutralization potency against the Omicron variant relative to the Delta variant in sera from previously infected patients, with particularly distinctive drops for patients who had an infection with the earlier Alpha and Delta variants.
Evolutionary processes at play
The findings reported in this study are congruent with other recent reports that support the notion of increased resistance of the Omicron variant to neutralization. This is contingent on either prior infection with some earlier variant or vaccination and actually more pronounced than in other variants of concern that has emerged over the course of the coronavirus disease 2019 (COVID-19) pandemic.
“The large number of mutations on the surface of the spike protein including the immunodominant RBD would be expected to help the virus escape antibodies elicited by vaccination or prior infection,” say study authors. “It is interesting that the Omicron variant evolved to retain its ability to bind ACE2 efficiently despite these extensive mutations”, they emphasize in this Science paper.
In any case, the currently ubiquitous Omicron variant appears to have evolved to selectively balance improved escape possibility from the neutralization process with its propensity to interact with the ACE2 receptor efficiently.
Finally, increasing antibody evasion and retaining strong interactions at the ACE2 interface are key factors contributing to the soaring transmission of the Omicron variant – with biological and important public health implications.