Are there any risk factors that predispose patients to severe COVID-19 outcomes? A recent study published in the medRxiv* preprint server sheds light on this popular question. Using a combination of population genetics to single-cell RNA sequencing, European researchers identified the respiratory epithelial cell-specific ELF5 gene as a potential mediator for severe COVID-19 cases.
Understanding the causes behind severe COVID-19 illness could prevent hospitalization and death. Scientists could then repurpose drugs that target and inhibit the activation of genes associated with severe illness for COVID-19 treatments.
Study details
The study used genetically informed plasma proteomics to find genes from over 2,375 potential protein targets that could explain the cause behind severe COVID-19 illness. They searched for a shared genetic architecture at protein-coding loci and narrowed their search to six unknown candidate causal proteins.
Increases in ELF5 gene expression elevates risk for poorer COVID-19 prognosis
One of six protein candidates, ELF5, stood out to the researchers. ELF5 was significantly and directly correlated with all four COVID-19 outcomes ranging from mild to severe illnesses. Additionally, there was evidence of a person’s genes predicting a person’s chances of requiring hospitalization for severe disease. Increases in the ELF5 gene led to 5-fold increased risk of developing severe COVID-19 illness.
The five other candidate proteins were also associated with a poorer COVID-19 prognosis, but not with as much firm evidence as to the ELF5 gene. For instance, G-CSF shared an estimated 64% genetic signal for more severe COVID-19 infection.
Genetic signal linked ELF5 expression to severe infection is specific to the lungs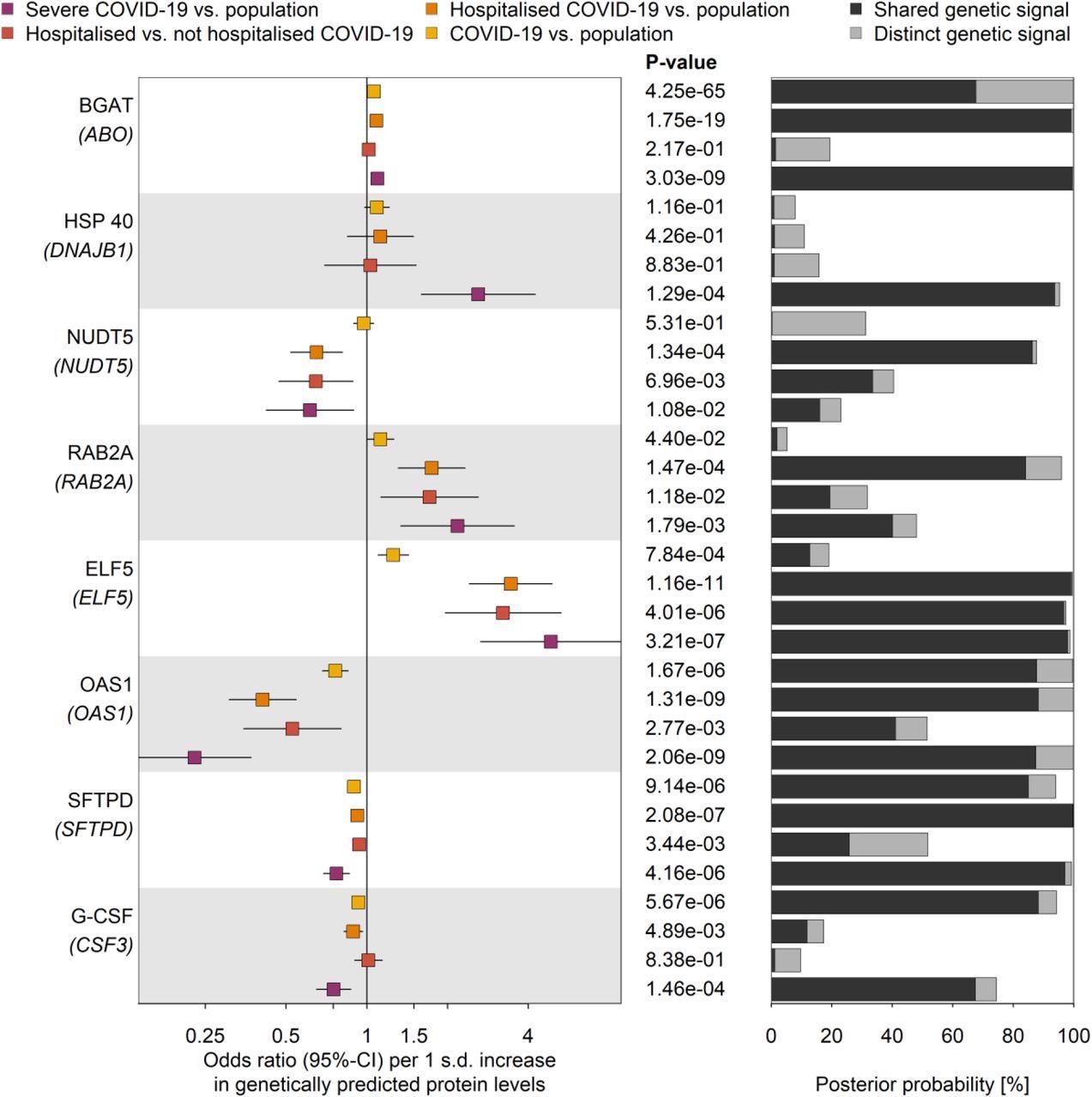
Proteins genetically linked to various COVID-19 outcomes. Odds ratios and 95%-CIs for the genetically predicted effect of protein levels on four different outcome definitions and control populations for COVID-19 (left), including protein targets with strong evidence for statistical colocalization for at least one definition (right). The column in the middle reports p-values. BGAT = Histo-blood group ABO system transferase; HSP 40 = DnaJ homolog subfamily B member 1; NUDT5 = ADP-sugar pyrophosphatase; RAB2A = Ras-related protein Rab-2A; ELF5 = ETS-related transcription factor Elf-5; OAS1 = 2’-5’-oligoadenylate synthase 1; SFTPD = Pulmonary surfactant-associated protein D; G-CSF = Granulocyte colony-stimulating factor
With a new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant known as rs61882275, there is an association between it and severe infection. However, the gene causing the severity remained unknown. Researchers in the current study identified the ELF5 gene as the leading causal gene behind severe SARS-CoV-2 infection.
The researchers then tested for colocalization of ELF5 gene expression across a variety of tissues that SARS-CoV-2 could infect. They found ELF5 is expressed in multiple tissues, but its expression in the lung was associated with a higher risk of severe COVID-19 infection and increased amounts of ELF5 in the blood.
ELF5 expression and a poor COVID-19 prognosis were specific to lung function.
ELF5 expression is found in different epithelial cells in the lungs
Using single nucleus RNA sequences across samples of healthy donors, the team found ELF5 is expressed in multiple types of respiratory epithelial cells. Sustentacular and Bowman gland cells from the olfactory mucosa and secretory epithelial cells from the nasopharynx and distal bronchioles of the lung had high ELF5 expression. These cells are also the ones that tend to be infected and express host proteins used by SARS-CoV-2, including ACE2 and TRMPRSS2.
When looking at the lungs of patients who died from COVID-19 infection, they found substantial alveolar damage consistent with the high ELF5 expression seen in secretory cells. However, the researchers note that AT2 but not AT1 lung cells maintain the same expression of ELF5. This corresponds with classical lineage markers for AT2 cells, such as SFTPD, and may suggest there are other candidate genes that interact with ELF5 gene expression.
Next, the researchers looked at ELF5 expression in the same tissue samples of two patients who died less than 2 weeks after becoming infected with SARS-CoV-2. Again, they observed a damaged olfactory epithelium, including injured Bowman glands and low ELF5 expression in cells. There were also injuries to the alveolar region of the lungs, with only a few AT2 cells remaining.
AT2 cells that exhibited high ELF5 expression formed clusters. In addition, secretory cells such as SCGB1A1+ and other epithelial cells of the airway mucosa showed ELF5 expression and were damaged during SARS-CoV-2 infection. With high ELF5 expression, there were also signs of remodeling in respiratory tissue samples, as there was a loss in the structural integrity in the lungs.
ELF5 expression was also found to be linked to severe COVID-19 illness because it is coexpressed with TMPRSS2 — a gene needed for viral entry into cells. There was a positive correlation between ELF5 and TMPRSS2 expression in sustentacular cells.
The results suggest ELF5 expression plays a role in the course of severe COVID-19 infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Pietzner M, et al. (2022) ELF5 is a respiratory epithelial cell-specific risk gene for severe COVID-19. medRxiv. Doi: https://doi.org/10.1101/2022.01.17.22269283, https://www.medrxiv.org/content/10.1101/2022.01.17.22269283v1
- Peer reviewed and published scientific report.
Pietzner, Maik, Robert Lorenz Chua, Eleanor Wheeler, Katharina Jechow, Julian D. S. Willett, Helena Radbruch, Saskia Trump, et al. 2022. “ELF5 Is a Potential Respiratory Epithelial Cell-Specific Risk Gene for Severe COVID-19.” Nature Communications 13 (1): 4484. https://doi.org/10.1038/s41467-022-31999-6. https://www.nature.com/articles/s41467-022-31999-6.