
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Viruses evade the host's innate immunity via suppressing the synthesis or activity of cytokines such as type I IFNs (IFN-α and IFN-β). Thus, IFN production is pivotal to antiviral immunity as a host defense mechanism. Deficiency of type I IFN response during the SARS-CoV-2 infection can result in life-threatening conditions. Several SARS-CoV-2 proteins, like non-structural protein 1 (NSP1), NSP3, NSP5, and ORF3a, inhibit interferon (IFN)-β1 transcription in the host. However, the mechanism by which SARS-CoV-2 specifically represses IFN-β1 messenger ribonucleic acid (mRNA) transcription is not yet established.
About the study
In the present study, the researchers explored the mechanism by which SARS-CoV-2 inhibits the synthesis of the inflammatory marker IFN-β.
The interaction between SARS-CoV-2 NSP2 and cellular Grb10-interacting GYF (glycine, tyrosine, phenylalanine) protein 2/4Eukaryotic homologous protein (GIGYF2/4EHP) complex was validated using the Proximity Ligation Assay (PLA). Further, NSP2 impact on the translational repression by GIGYF2 was determined using the λN-BoxB tethering approach.
GIGYF2 tethering experiments were conducted in SARS-CoV-2 wild-type (WT), GIGYF2-KO, and 4EHP-KO cells with or without ectopic 4EHP expression to determine the impact of 4EHP on NSP2-induced GIGYF2-mediated translational repression.
The role of GIGYF2 in the regulation of IFN-β synthesis was examined in WT, GIGYF2-KO, and 4EHP-KO HEK293 cells and in two lung epithelial cell lines A549 and Calu-3. Later, the GIGYF2/4EHP complex's role in the repression of IFN-β production was also determined.
To determine whether IFN-β1 mRNA 3' untranslated region (3' UTR) exerts its silencing effect through GIGYF2, a Renilla luciferase reporter (R-Luc) fused to the Ifnb1 3' UTR was transfected into WT, GIGYF1-KO, GIGYF2-KO, or 4EHP-KO cells.
The role of GIGYF2 in the antiviral immune response to RNA viruses was assessed using a green fluorescent protein (GFP)-tagged mutant variant (VSVΔ51) of the vesicular stomatitis virus (VSV). Whether GIGYF2 directly targets virus-induced activation of signaling pathways upstream and downstream of IFN-β was assessed by GIGYF2-KO cells co-transfected with a Firefly luciferase reporter (F-Luc).
The role of NSP2 in regulating IFN expression was determined using the SARS-CoV-2 NSP1, NSP2, or Envelope (E) protein expressed in HEK293 cells co-transfected with the R-Luc reporter fused to the Ifnb1 3' UTR. Further, NSP2's role in the IFN expression of lung epithelial cell line A549 was determined using lentiviral vectors.
Results
The results indicated that SARS-CoV-2 through NSP2 enhances the GIGYF2/4EHP complex-mediated inhibition of the IFN-β1 mRNA translation.
NSP2 specifically increases the translational repression by GIGYF2 compared to that of GIGYF1. NSP2 interacts directly with the GIGYF2 protein in humans and not with 4EHP. Further, NSP2 promotes GIGYF2-induced translational repression in a 4EHP-dependent manner and enhances the interaction of GIGYF2 and 4EHP. Although GIGYF2 and 4EHP suppress IFN-β production, more potent repression was exhibited by GIGYF2 than by 4EHP. The formation of the GIGYF2/4EHP complex has a pivotal role in the repression of IFN-β synthesis.
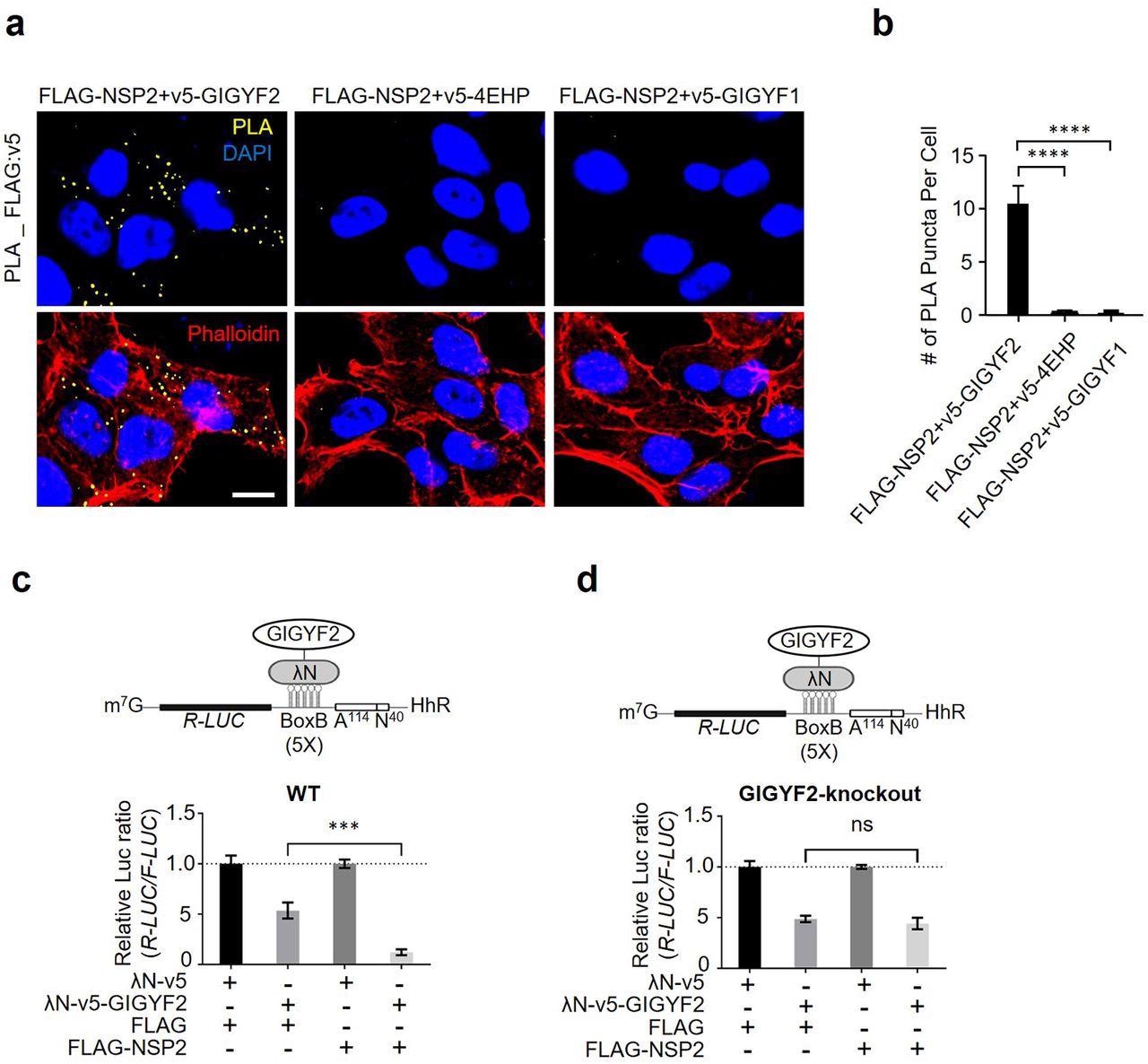
NSP2 bolsters the GIGYF2/4EHP translational repression complex. (a) PLA detection of NSP2-GIGYF2 interactions visible as fluorescent punctate in HEK293T cells transfected with vectors expressing v5-tagged GIGYF2, 4EHP, or GIGYF1 together with FLAG-NSP2. 24 h post-transfection cells were fixed and subjected to PLA using FLAG and v5 antibodies. PLA signals are shown in yellow. Nucleus and actin cytoskeleton were counterstained with DAPI (blue) and phalloidin (red), respectively. Scale bar= 10 µm. (b) Quantification of positive PLA signals in (a). The number of PLA signals from at least 30 cells was counted in each sample. ****P<0.0001 one-way ANOVA with Bonferroni’s post hoc test; n=5 independent replicates. Data are presented as mean ± SD. (c) WT HEK293 cells were co-transfected with plasmids expressing either λN-v5-GIGYF2 or λN-v5 as control, along with R-Luc-5BoxB-A114-N40-HhR and F-Luc (as control), followed by dual-luciferase measurement assay. Data are presented as mean ± SD (n=3). ***p<0.001, one-way ANOVA with Bonferroni’s post hoc test. The schematic shows a graphic model of λN-v5-GIGYF2 tethering system. (d) Analysis of λN-v5-GIGYF2 tethering-induced silencing in GIGYF2-KO cells that overexpress FLAG-NSP2. Data are presented as mean ± SD (n=3). ns= non-significant, one-way ANOVA with Bonferroni’s post hoc test.
Another RNA virus, VSV, also suppresses IFN-β synthesis via GIGYF2/4EHP complex. The findings further demonstrated that the GIGYF2/4EHP complex-mediated antiviral immune response via direct repression of IFN-β1 mRNA translation and not by directly affecting the signaling pathways upstream or downstream of IFN-β or RNA virus sensors.
Additionally, SARS-CoV-2 NSPs like NSP1 and NSP14 are also involved in the dysregulation of the host mRNA translation. NSP1 inhibits host mRNA ribosomal entry site but promotes SARS-CoV-2 mRNA translation. Hence, NSP1 through nonspecific inhibition of host mRNAs such as IFN-β1 translation results in the depletion of labile antiviral factors such as STAT254 and Tyk2. Further, NSP14 blocks global mRNA translation and possibly inhibits the expression of IFN-stimulated genes (ISGs).
The structural analysis of SARS-CoV-2 GIGYF2 revealed a previously uncharacterized GIGYF2 region (744-940), a singular helix alpha called GIGYF2 long helix region (GIGYF2-LHR). In addition, the docking simulations gave rise to models that demonstrate the NSP2 and GIGYF2/4EHP complex interactions consistent with the present structural analyses of these proteins and the literature data on affinity purification-mass spectrometry (AP-MS) assay.
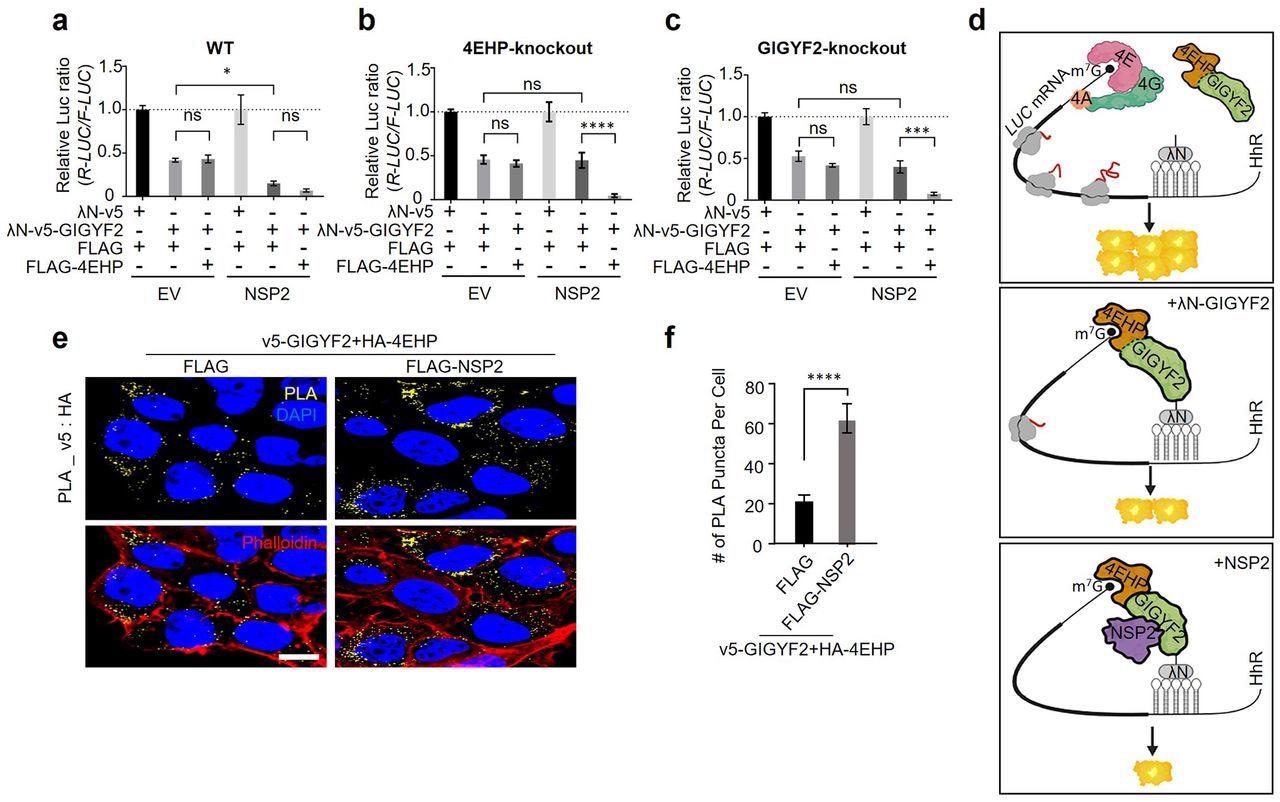
NSP2 augments GIGYF2/4EHP complex-mediated translational repression by enhancing the GIGYF2 interaction with 4EHP. (a) WT cells were co-transfected with vectors expressing either λN-v5-GIGYF2 or λN-v5 control, along with R-Luc-5BoxB-A114-N40-HhR and F-Luc (as control), in combination with FLAG-4EHP or FLAG-Empty plasmids. Dual-luciferase assay was performed 24 h post-transection. Data are presented as mean ± SD (n=3). *p<0.05, one-way ANOVA with Bonferroni’s post hoc test. (b-c) GIGYF2-tethering assay carried out in 4EHP-KO cells in (b) and GIGYF2-KO cells in (c). Data are presented as mean ± SD (n=3). ns= non-significant, **p<0.01, ***p<0.001, one-way ANOVA with Bonferroni’s post hoc test. (d) Graphic illustration of the GIGYF2/4EHP-mediated induction of translational repression by NSP2. (e) PLA assay for detection of GIGYF2-4EHP interactions in HEK293T cells transfected with vectors expressing v5-GIGYF2 and HA-4EHP together with FLAG-Empty or FLAG-NSP2. Cells were fixed and subjected to PLA using v5 and HA antibodies 24 h post-transfection. Scale bar= 10 µm. (f) Quantification of positive PLA signals from (e). The number of PLA signals from at least 20 cells was counted in each sample. n=5 independent experiments. Data are presented as mean ± SD (n=5). ****P< 0.0001, one-way ANOVA with Bonferroni’s post hoc test
Conclusions
The study findings demonstrated that SARS-CoV-2 encoded NSP2 co-opts the cellular GIGYF2 protein and thereby increases GIGYF2 affinity towards the mRNA cap-binding protein 4EHP, resulting in suppressing the synthesis of the main immunostimulatory cytokine IFN-β1. Thus, during SARS-CoV-2 infection, the depletion of 4EHP or GIGYF2 will substantially elevate the IFN-β production, resulting in a reduction in the viral infection.
Further, the three-dimensional structure of LHR and the NSP2-GIGYF2/4EHP interaction will be beneficial in understanding the molecular basis of the proposed NSP2/GIGYF2/4EHP complex. Most importantly, the identification of GIGYF2-LHR has immense significance during the development of recombinant small molecules or peptides to inhibit the abolish the interaction of NSP2 with GIGYF2.
Overall, the study findings presented a new target for protecting the antiviral innate immune response towards RNA viruses, including SARS-CoV-2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
SARS-CoV-2 impairs interferon production via NSP2-induced repression of mRNA translation, Jung-Hyun Choi, Xu Zhang, Christine Zhang, David L. Dai, Jun Luo, Reese Ladak, Qian Li, Shane Wiebe, Alex C.H. Liu, Xiaozhuo Ran, Jiaqi Yang, Parisa Naeli, Aitor Garzia, Lele Zhou, Niaz Mahmood, Qiyun Deng, Mohamed Elaish, Rongtuan Lin, Tom Hobman, Jerry Pelletier, Tommy Alain, Silvia Vidal, Thomas Duchaine, Mohammad T. Mazhab-Jafari, Xiaojuan Mao, Seyed Mehdi Jafarnejad, Nahum Sonenberg, bioRxiv, 2022, doi: https://doi.org/10.1101/2022.01.19.476693, https://www.biorxiv.org/content/10.1101/2022.01.19.476693v1
- Peer reviewed and published scientific report.
Xu, Zhang, Jung-Hyun Choi, David L. Dai, Jun Luo, Reese Jalal Ladak, Qian Li, Yimeng Wang, et al. 2022. “SARS-CoV-2 Impairs Interferon Production via NSP2-Induced Repression of MRNA Translation.” Proceedings of the National Academy of Sciences 119 (32). https://doi.org/10.1073/pnas.2204539119. https://www.pnas.org/doi/full/10.1073/pnas.2204539119.