To date, over 413 million confirmed cases of coronavirus disease 2019 (COVID-19), including more than 5.82 million deaths, have been reported globally. Nine different vaccines have been approved or authorized for emergency use to curb the pandemic; however, the waning of vaccine efficacy suggested by several reports necessitates the development of a therapeutic approach against SARS-CoV-2.
Study: N-acylethanolamine acid amide hydrolase is a novel target for drugs against SARS-CoV-2 and Zika virus. Image Credit: nechaevkon / Shutterstock

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
About the study
The present study assessed the impact of NAAA inhibitors on the replication process of viruses, like SARS-CoV-2 and ZIKV, that utilize lipid droplets to facilitate their replication.
A549 (a lung cell line) and Huh-7 (a liver cell line) were treated with 10% fetal bovine serum (FBS), 100 µg/mL streptomycin, and 100 U/mL penicillin. These cells were transfected by nucleoporation, single-celled clones were isolated, and sequential analysis of polymerase chain reaction (PCR) was conducted to select the edited clones. The cells were infected with ZIKV or SARS-CoV-2 and coxsackievirus B5 (COXB5).
Cells infected with ZIKV or SARS-CoV-2 were assessed by flow cytometry, while the COXB5-infected cells were examined by Western blot analysis. Total ribonucleic acid (RNA) of the infected cells was extracted, reverse-transcribed as complementary DNA (cDNA), and amplified by reverse transcription-polymerase chain reaction (RT-PCR).
NAAA messenger RNA (mRNA) was measured before and after both A549 and Huh-7 cells were infected with ZIKV. Total RNA was collected 24 hours and 48 hours after the infection and the upregulation of NAAA was confirmed by treating the A549 cells with both anti-ZIKV and anti-NAAA antibodies. The study also explored the localization of the NAAA and ZIKV anti-flavivirus NS1 protein.
The antiviral potency of a highly potent NAAA inhibitor ARN077 was tested by administering the drug 15 minutes before infecting the cells with ZIKV at concentrations of 0.3, 1, 3, and 10 µM. The potency of the drug was also tested on COXB5-infected cells.
Results
The study showed that NAAA expression increased by almost two and three times in A549 and Huh-7 at 48 hours, respectively, while an insignificant difference was observed at 24 hours. The upregulation of NAAA expression was detected only in ZIKV-infected cells. Also, NAAA and ZIKV NS1 proteins were observed in approximately 30% of the cytoplasmic vesicles.
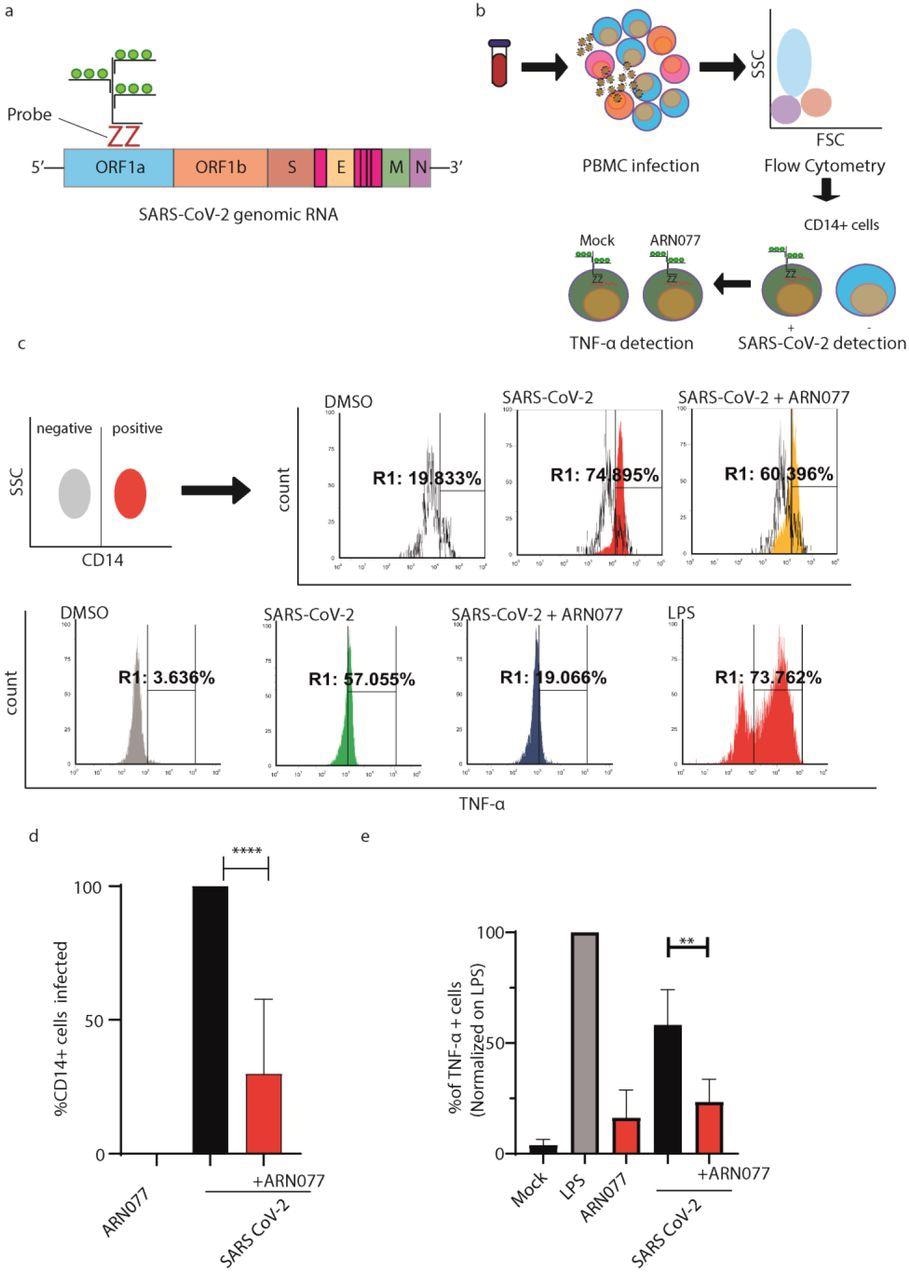
ARN077 reduces SARS-CoV-2 infection and controls inflammation in human PBMCs. a) Schematic illustration on the design of SARS-CoV-2 genomic probe, which binds a conserved region of ORF1a. b) Experimental workflow of primeflow assay combined with anti-CD14 staining. Briefly, PBMC were infected with SARS-CoV-2, treated or not with ARN077 (30 μM). Then, cells were treated with Brefeldin-α prior fixation and processed by Flow Cytometry. PBMCs were then analyzed for SARS-CoV-2 genomic and TNF-α content. c) Representative analysis of PBMC derived macrophages infected or not with SARS-CoV-2. Briefly, CD14+ cells (monocytes and macrophages) were identified by immunostaining. Then, the CD14+ cell population was screened for SARS-CoV-2 intracellular genomes and TNF-α production. DMSO was used as mock-treated control. d) Statistical analysis on SARS-CoV-2 genomic content on CD14+ cells, treated or not with ARN077. Data are expressed as mean ± SD and analyzed with One-Way Anova ( **** p<0,0001, n=8, α=0,5) e) Statistical analysis performed on % of TNF-α+ cells, treated or not with ARN077 and infected or not with SARS-CoV-2. Data were normalized on cells treated with Lipopolysaccharide (LPS). Data are expressed as mean ± SD and analyzed with One-Way Anova (** p<0,01, n=8, α=0,5).
A five-fold decrease in the amount of ZIKV plaque-forming units (PFU) was found in A549 NAAA cells. The Western blot analysis confirmed a reduction of 20-fold in viral content. A dose-dependent reduction of ZIKV protein and ZIKV genomes was observed with ARN077 drug treatment. In the case of COXB5, NAAA was inefficient in reducing viral replication, while the release of SARS-CoV-2 decreased by five-fold by NAAA treatment.
In comparing antiviral activities of NAAA inhibitors versus NAAA ablation, ARN077 caused a four-fold reduction in released SARS-CoV-2 virions. Furthermore, NAAA ablation reduced the replication of both SARS-CoV-2 Delta (B.1.617.2) and Alpha (B.1.17) variants of concern (VOCs). Also, SARS-CoV-2 caused a remarkable decrease in transcripts of peroxisome proliferator receptor-α (PPAR-α) while NAAA maintained high levels of the same. Therefore, PPAR-α levels were notably low in SARS-CoV-2-infected cells and significantly higher in cells infected with NAAA.
The presence of ARN077 led to a macrophage-induced 70% reduction of SARS-CoV-2 genomes. Tumor necrosis factor-alpha (TNF-α) production triggered by SARS-CoV-2 was also eliminated by ARN077 as compared to cells not treated with the drug.
The number of vacuoles containing the autophagic marker of microtubule-associated protein light chain 3 (LC3) was reduced in SARS-CoV-2-infected cells, while the presence of NAAA prevented this decrease. Also, SARS-CoV-2 decreased the compartmentalization of LC3 inside the vacuoles as compared to that in the cytosol. In contrast, NAAA-infected cells showed higher compartmentalization of LC3 in the vacuoles versus the cytosol. This indicated that the increased compartmentalization of LC3 induced by NAAA in vacuoles effectively reduced the LC3 compartments generated by the virus.
Conclusion
The study findings concluded that NAAA reduced SARS-CoV-2 and ZIKV replication by approximately five times. NAAA treatment effectively reduced viral replication and the release of viral lipid drops, while NAAA autophagy improved viral clearance. Since NAAA is an autophagy-related protein-coding enzyme, it cannot inhibit viruses like COXB5, which replicate using autophagy-independent intracellular membranes.
The researchers believe that exploring the impact of the NAAA enzyme on viral replication reveals new approaches via which SARS-CoV-2 and ZIKV can affect human immune responses. This knowledge can facilitate the development of a new antiviral therapy to combat the replication of the two viruses and curb disease severity.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
N-acylethanolamine acid amide hydrolase is a novel target for drugs against SARS-CoV-2 and Zika virus, Michele Lai, Veronica La Rocca, Rachele Amato, Elena Iacono, Carolina Filipponi, Elisa Catelli, Lucia Bogani, Rossella Fonnesu, Giulia Lottini, Alessandro De Carli, Alessandro Mengozzi, Stefano Masi, Paola Quaranta, Pietro Giorgio Spezia, Giulia Freer, Paola Lenzi, Francesco Fornai, Daniele Piomelli, Mauro Pistello, bioRxiv 2022.02.08.479661, doi: https://doi.org/10.1101/2022.02.08.479661, https://www.biorxiv.org/content/10.1101/2022.02.08.479661v1