
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Several variants of SARS-CoV-2 have been detected to date following its emergence in late 2019. However, the SARS-CoV-2 Omicron variant is currently the predominant variant of concern (VOC) due to its high transmissibility and the ability to escape from natural- and vaccine-induced immunity.
Three subvariants of SARS-CoV-2 Omicron have been identified so far, namely BA.3, 21L/BA.2, and 21K/BA.1. In contrast to the 21K/BA.1 Omicron variant with a rapid transmission rate even in highly vaccinated populations, 21L/BA.2 was observed less frequently in most countries worldwide. However, recently in Denmark, the 21L/BA.2 Omicron variant has become predominant over the SARS-CoV-2 21K/BA.1 variant, suggesting a possibility of epidemiological change of 21L/BA.2 in other nations.
About the study
In the current study, the researchers characterized the initial cases of the SARS-CoV-2 21L/BA.2 Omicron variant in Marseille, France.
The nasopharyngeal samples procured from patients of the University Hospital Institute Méditerranée Infection was tested for SARS-CoV-2 infection using the reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Further, SARS-CoV-2 variants-specific qPCR assays identifying the spike (S) mutations E484K, K417N, L452R, or P681H merged with TaqPath coronavirus disease 2019 (COVID-19) kit targeting the SARS-CoV-2 ORF1, nucleocapsid (N), and S genes were conducted as per the French recommendations.
Amino acid and nucleotide changes in the 21L/BA.2 Omicron genomes compared to the genome of SARS-CoV-2 Wuhan-Hu-1 isolate were evaluated using the Nextclade tool. The genomic identification of 21L/BA.2 was performed via next-generation sequencing with the Oxford Nanopore Technology (ONT). Pangolin lineages and Nextstrain clades were determined using the Pangolin and Nextclade web applications.
Further, phylogeny was reconstructed and visualized using the Nextstrain/new CoV (nCoV) tool and Auspice, respectively. Genomes that were genetically closest to the 21L/BA.2 Omicron was determined using the Usher and global initiative for sharing all influenza data (GISAID) BLAST tools.
Findings
The results indicate that 13 SAR-CoV-2 21L/BA.2 Omicron cases were identified from Marseille between November 28, 2021, to January 31, 2022, using variant-specific qPCR and next-generation sequencing. The initial two cases were detected in two couples who traveled back from Tanzania. These travelers were tested positive following three weeks of receiving the third dose of the SARS-CoV-2 Pfizer-BioNTech vaccine.
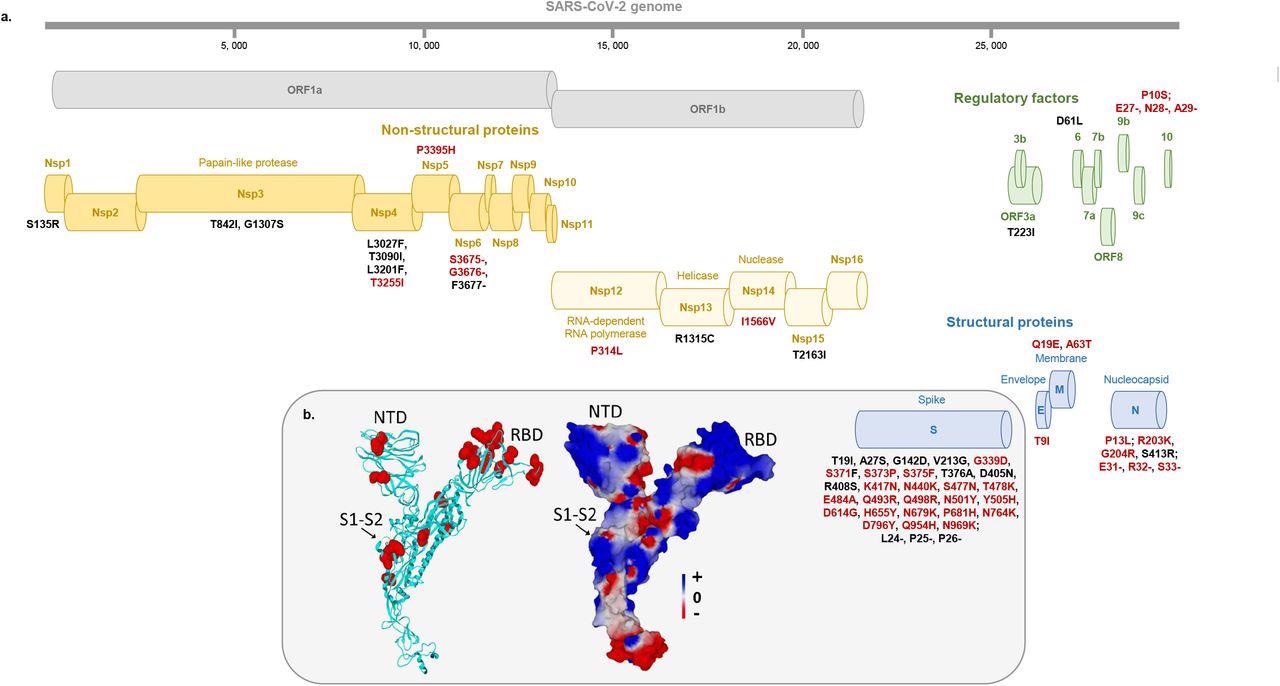
Map of the Omicron 21L/BA.2 spike protein with signature amino acid substitutions and deletions (a) and structural features of 21L/BA.2 Omicron variant spike protein (b) a: Amino acid substitutions and deletions shared with the 21K/BA.1 Omicron variant are indicated by a red font. b: Structural model of the Omicron 21L/BA.2 spike protein with mutations highlighted in red atomic spheres (left panel) or in electrostatic surface rendering (right panel). Note the flat surface of the N-terminal domain that faces lipid rafts of the host cell membrane. The S1-S2 cleavage site is indicated by an arrow. The color scale for the electrostatic surface potential (negative in red, positive in blue, neutral in white) is indicated. NTD, N-terminal domain; RBD, Receptor binding domain.
The third case had a history of contact with a COVID-19-positive patient and migrant patients with anonymous status of SARS-CoV-2 and received the third dose of Pfizer-BioNTech vaccine seven weeks before the 21L/BA.2 Omicron diagnosis. Finally, two other 21L/BA.2 patients originated from the United Kingdom (UK) and the Netherlands. Information about the remaining eight 21L/BA.2 Omicron-positive patients were unavailable.
Every 21L/BA.2-positive respiratory samples demonstrated the same S mutation combinations in real-time qPCR, which was negativity for E484K, P681R, and L452R, and positivity for K417N. Further, the TaqPath COVID-19 kit showed positive signals for all three S, N, and ORF1 genes targeted.
The 21L/BA.2 Omicron variant genomes demonstrated a mean of 65.9±2.5 nucleotide substitutions, 31.0±8.3 nucleotide deletions, 49.6±2.2 amino acid substitutions, and 12.4±1.1 amino acid deletions.
Phylogeny indicated that the nine 21L/BA.2 genomes detected in the current study were associated with three distinct clusters of 21L/BA.2 Omicron variant genomes from Singapore and Nepal, France and Denmark, England and South Africa.
The 21L/BA.2 Omicron variant had a substantially enlarged and flattened N-terminal domain (NTD) surface compared to 21K/BA.1 in the molecular modeling and structural predictions, probably due to the absence of 143-145 nucleotide deletions in the 21L/BA.2. This indicated the advanced 21L/BA.2 Omicron adaptability to the electronegative surface of lipid rafts relative to the 21K/BA.1 Omicron variant.
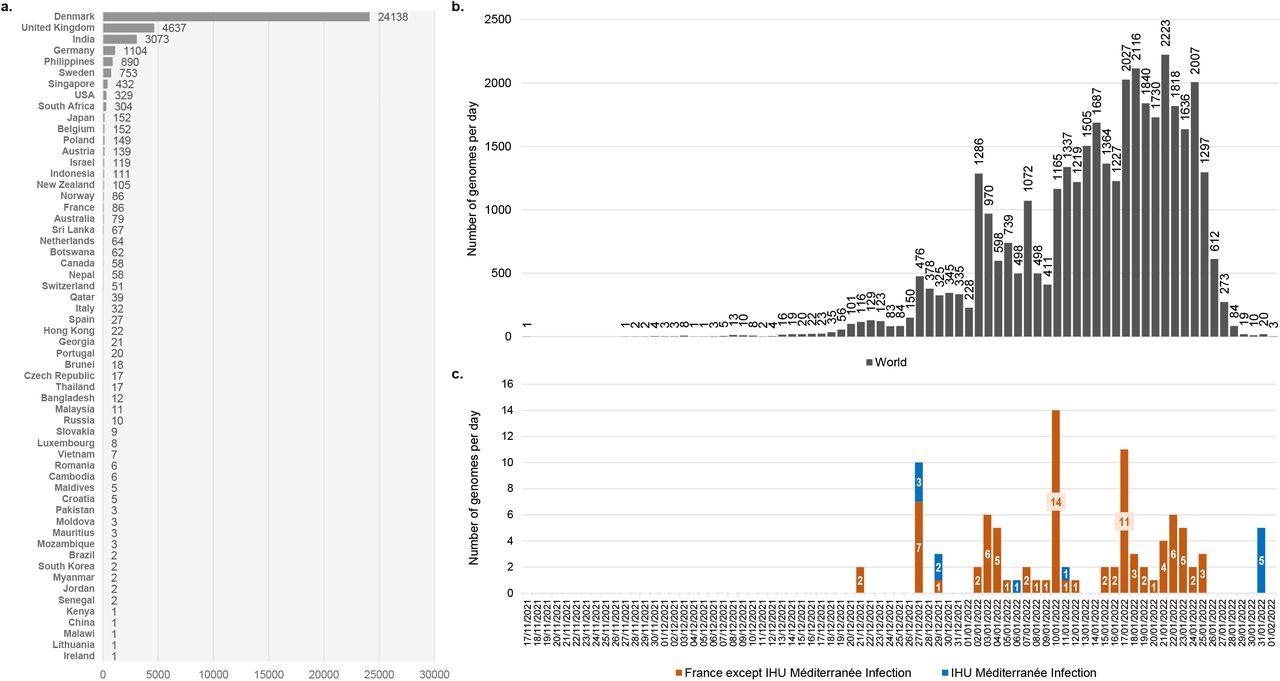
Number of genomes of the SARS-COV-2 21L/BA.2 Omicron variant available in GISAID and chronology of collections of respiratory samples from where they were obtained a: Number of genomes of the SARS-COV-2 21L/BA.2 Omicron variant available in the GISAID sequence database (https://gisaid.org/)15 as of 02/02/2022. b: Chronology of SARS-CoV-2 diagnoses with the 21L/BA.2 Omicron variant for genomes deposited in the GISAID sequence database and obtained worldwide. c: Chronology of SARS-CoV-2 diagnoses with the 21L/BA.2 Omicron variant for genomes deposited in the GISAID sequence database and obtained in France or in our university hospital institute. The number of genomes was analyzed until 02/02/2022. Total number of genomes analyzed was 36,428. A total of 1,093 genomes were excluded as the date of sample collection was incomplete (days or months were lacking).
Conclusions
The present study highlights the need for 1) close surveillance at the center, country, and global levels to monitor the occurrence and clinical outcome of the SARS-CoV-2 21L/BA.2 Omicron variant as no study is available to date describing the frequency of severe, mild, and asymptomatic forms of COVID-19 associated with 21L/BA.2; 2) phenotypic assessment through inoculation on permissive cells to determine the susceptibility of the emerging SARS-CoV-2 variants to neutralization by the previous COVID-19 infection- or vaccine-induced anti-S antibodies; and 3) the real-time close genomic and molecular surveillance of the emergence, transmission, and disappearance of SARS-CoV-2 variants to predict the emergence and outcomes of novel SARS-CoV-2 variants.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
First cases of infection with the 21L/BA.2 Omicron variant in Marseille, France, Philippe Colson, Jérémy Delerce, Mamadou Beye, Anthony Levasseur, Céline Boschi, Linda Houhamdi, Hervé Tissot-Dupont, Nouara Yahi, Matthieu Million, Bernard La Scola, Jacques Fantini, Didier Raoult, Pierre-Edouard Fournier, medRxiv 2022.02.08.22270495; doi: https://doi.org/10.1101/2022.02.08.22270495, https://www.medrxiv.org/content/10.1101/2022.02.08.22270495v1
- Peer reviewed and published scientific report.
Colson, Philippe, Jérémy Delerce, Mamadou Beye, Anthony Levasseur, Céline Boschi, Linda Houhamdi, Hervé Tissot‐Dupont, et al. 2022. “First Cases of Infection with the 21L/BA.2 Omicron Variant in Marseille, France.” Journal of Medical Virology 94 (7): 3421–30. https://doi.org/10.1002/jmv.27695. https://onlinelibrary.wiley.com/doi/10.1002/jmv.27695.