SARS-CoV-2 infection affects the vasculature as indicated by endothelial cell activation, thrombosis, endothelitis, and vascular permeability in severe coronavirus disease 2019 (COVID-19) patients during the acute stage.
The current research was conducted to provide molecular insights into the role of the endothelium in SARS-CoV-2 infection, the importance of miRNA as pathway and disease-specific biomarkers, and the potential of barrier stabilizing treatment to combat COVID-19.
Study: Cardiovascular Signatures of COVID-19 Predict Mortality and Identify Barrier Stabilizing Therapies. Image Credit: Kateryna Kon/Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Study Design
In the study, 241 patients with community-acquired acute SARS-CoV-2 infection between May 2020 and December 2020 were enrolled in two quaternary-care hospitals in Toronto, Canada. The estimation of various inflammatory, cardiac, and endothelial dysfunction markers and miRNA atlas development in these patients were performed, and the prognostic value was assessed by random forest model machine learning approach.
The amalgamation of the electronic health data records with multi-omics datasets by machine learning in this study provided a better metric for mortality estimation due to severe COVID-19.
miRNA pathway analysis was performed using Kyoto encyclopedia of genes and genomes (KEGG)/Biokarta/Reactome databases and tested for enrichment by a hypergeometric test.
Findings
The researchers observed a higher mortality risk due to COVID-19 in patients with severe disease compared to patients with less severe phenotypes. Moreover, within admitted patients, only previous history of coronary artery disease and age at the time of hospital admission was associated with mortality.
During hospitalization, 44.2% of COVID-19 patients were admitted to the ICU, with 30.3% and 69.7% required non-invasive and invasive ventilation, respectively, and 27.6% were subjected to extracorporeal membrane oxygenation.
Compared to the SARS-CoV-2 severity matched control group, COVID-19 patients of severe category stayed in the ICU for a longer duration (mean 13 days vs mean 8 days), displayed acute respiratory distress syndrome (ARDS) and suffered worse oxygenation, and required a higher fraction of inspired oxygen (FiO2) and lower partial pressure of alveolar oxygen (PaO2)/FiO2.
The researchers observed that among the different proinflammatory markers, a significant difference existed between angiopoietin-2 (Ang-2) and myeloperoxidase (MPO) in SARS-CoV-2 and mild SARS-CoV-2-negative patients.
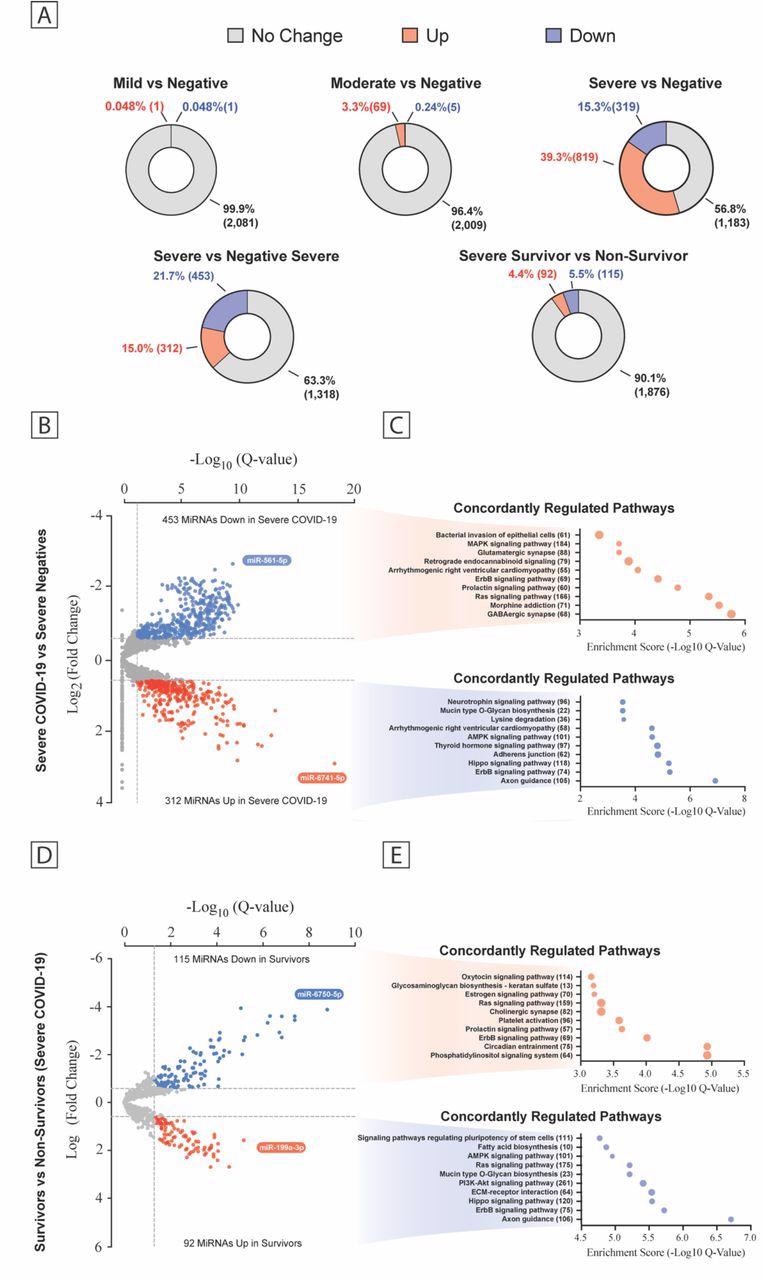
Plasma MiRNA Transcriptome Across the COVID-19 Severity. (a) Pie chart percent modulation of the transcriptome for all subgroups studied. Volcano plots of differentially expressed miRNA between patient groups (b, d) with predicted KEGG terms (with enrichment score below and number of genes to the right) for pathways of deregulated miRNAs shown beside each corresponding region of the volcano plot (c, e). Data are displayed as false discovery rate (FDR) adjusted P values (Q values) vs the log2 fold change, with dashed lines drawn to define restriction boundaries
There were differences in severity within the SARS-CoV-2-positive cohort of nine biomarkers [Ang-2, Interleukin-6 (IL-6), IL-8, soluble intercellular adhesion molecule-1 (sICAM-1), soluble vascular cell adhesion molecule-1 (sVCAM-1), endothelin-1 (ET-1), soluble triggering receptor expressed on myeloid cells-1 (sTREM-1), soluble E-selectin (sE-Selectin), and MPO] but no significant difference was demonstrated in critically ill SARS-CoV-2 positive or -negative patients.
Univariable analysis revealed that among all admitted SARS-CoV-2-positive patients, a high concentration of Ang-2 was associated with higher mortality, while in severe COVID-19 patients both Ang-2 and sVCAM-1 was associated with mortality.
In COVID-19 non-survivors, significantly higher concentrations of IL-6, Ang-2, and MPO were observed. However, over time in COVID-19-positive and -negative patients with severe illness, only MPO and IL-6 remained significantly different.
While comparing severe COVID-19-positive and -negative patients by miRNA sequencing analysis, the researchers observed the presence of 765 differentially expressed miRNAs. There was broad enrichment of pathways such as arrhythmogenic right ventricular cardiomyopathy, Erb-B2 receptor tyrosine kinase 2 (ErbB2) signaling, and adherens junctions.
In severe COVID-19 survivors and COVID-19 non-survivors, the expression of 207 miRNA, including the enrichment pathway for extracellular matrix-receptor interactions, platelet activation, ErbB2, and rat sarcoma (Ras), was observed.
At the time of admission, using only available clinical features a low area under the receiver operating characteristic (AUROC) was observed [AUROC 0.44, 95% confidence interval (CI)]. The incorporation of either protein expression or miRNA data improved the performance of AUROC to 0.82 and 0.76, respectively. As per the machine learning model, TREM-1, MPO, and Ang-2 were the top contributing features before any other clinical factors.
The team found that miRNAs constituted the highest-ranking factors compared to other clinical metrics with miRNAs such as miRs-30b/c/e, 181a-5p, -6080, -339p, and -199a-3p, highly specific for COVID-19-related severity and mortality, while no significant association with mortality was observed in the case of miR-1 (micro RNA-1).
The researchers observed significant induction of endothelial barrier dysfunction in moderate to severe COVID-19 patients compared to mild negative patients. The permeability of endothelial cells (EC) was well correlated with the levels of Ang-2, IL-6, IL-8, high-sensitivity cardiac troponin (hs-cTnI), sTREM-1, and ET-1.
Gene enrichment analysis revealed that in severe to moderate COVID-19 patients, there was an altered expression of endothelial genes related to acute inflammatory response and histone decyclase activity while in mild patients, alteration was observed in genes related to priming of antiviral responses. In the moderate to the severe group, pathways were broadly related to cell motility, cell stress responses, developmental processes, reorganization of cell structure, and actin mobilization.
The researchers observed that in ECs exposed to plasma of severe and moderate COVID-19 patients, there was significant disruption in the expression of vascular endothelial (VE)-cadherin and Claudin-5 and localization of junction. Further co-treatment with vascular stabilizing drugs like Q-peptide or Slit2-N prevented EC barrier disruption and permeability. Nangibotide and dexamethasone maintained junctional protein expression but only in specific settings.
Conclusion
This study showed that in severe SARS-CoV-2 patients, multiple biomarkers involved in inflammatory and EC activation pathways were associated with mortality due to COVID-19.
The observations obtained by plasma microRNA atlas generation in this study revealed that the exposure of COVID-19 plasma with EC resulted in gene expression responses and barrier dysfunction of EC, which were severity-specific and mitigated by using Q- Peptide and Slit2-N. Taken together, the findings of this study highlighted the need for exploring novel vascular stabilizing therapies for the treatment of pathologies involving endothelial dysfunction in COVID-19 patients.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Cardiovascular Signatures of COVID-19 Predict Mortality and Identify Barrier Stabilizing Therapies. Dakota Gustafson, Michelle Ngai, Ruilin Wu, Huayun Hou, Alice Schoffel, Clara Erice, Serena Mandla, Filio Billia, Michael D. Wilson, Milica Radisic, Eddy Fan, Uriel Trahtemberg, Andrew Baker, Chris McIntosh, Chun-Po S. Fan, Claudia C. dos Santos, Kevin C. Kain, Kate Hanneman, Paaladinesh Thavendiranathan, Jason E. Fish, Kathryn L. Howe, medRxiv, 2022.02.08.22270636, https://doi.org/10.1101/2022.02.08.22270636 https://www.medrxiv.org/content/10.1101/2022.02.08.22270636v1
- Peer reviewed and published scientific report.
Gustafson, Dakota, Michelle Ngai, Ruilin Wu, Huayun Hou, Alice Carvalhal Schoffel, Clara Erice, Serena Mandla, et al. 2022. “Cardiovascular Signatures of COVID-19 Predict Mortality and Identify Barrier Stabilizing Therapies.” EBioMedicine 78 (April): 103982. https://doi.org/10.1016/j.ebiom.2022.103982. https://www.thelancet.com/journals/ebiom/article/PIIS2352-3964(22)00166-9.