
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Globally, several World Health Organization (WHO)-approved coronavirus disease 2019 (COVID-19) vaccines are available that are highly potent in preventing COVID-19. Simultaneously, new SARS-CoV-2 VOCs continue to emerge, primarily in countries where vaccine coverage is low. Recently, SARS-CoV-2's new VOC Omicron post-emerging in South Africa, being much more aggressive than its predecessor Delta variant, spread worldwide quickly. Additionally, studies have evidenced that it caused incessant breakthrough infections (BTIs) even among vaccinated individuals globally.
Moreover, even if a vaccinated individual has a favorable prognosis and significantly reduced COVID-19-related morbidity and mortality, they may become a possible viral carrier and trigger viral dissemination within the community.
About the study
In the present study, researchers screened 29 fully vaccinated (with two vaccine doses) individuals irrespective of their age and gender in the state of Bahia who contracted reinfection and remained asymptomatic or developed symptoms during the course of the disease.
They gathered the oropharyngeal and nasal swabs of all the 29 study participants at the Laboratory of Virology at the Federal University of Bahia. There were also four 7- and 11-year-olds who were unvaccinated.
They detected SARS-CoV-2 using a quantitative reverse transcriptase-polymerase chain reaction (RT-qPCR) assay. The researchers filtered out samples with cycle threshold (Ct) values of ≤30 and subjected those to viral genomic sequencing using Nanopore technology. Running negative control samples helped filter out any possible sample contamination with less than 2% mean coverage.
The researchers performed all the study experiments in a biosafety level-2 (BSL-2) cabinet. They used a superscript IV reverse transcriptase kit for complementary DNA (cDNA) synthesis; subsequently, cDNA was subjected to multiplex polymerase chain reaction (PCR) sequencing using the Q5 High Fidelity Hot-Start DNA Polymerase and a set of specific primers.
The newly identified SARS-CoV-2 isolates were assigned a lineage using the Pangolin-lineage classification software. Subsequently, these isolates were mapped to a pool of 3,441 genomes, collected worldwide up to October 28th, 2021, to draw phylogenetic inferences.
Study findings
The self-reported COVID-19 symptoms of the study participants ranged from asymptomatic to mild, and their RT-qPCR Ct values varied between 14.6 and 27.2, even in individuals who declared themselves asymptomatic.
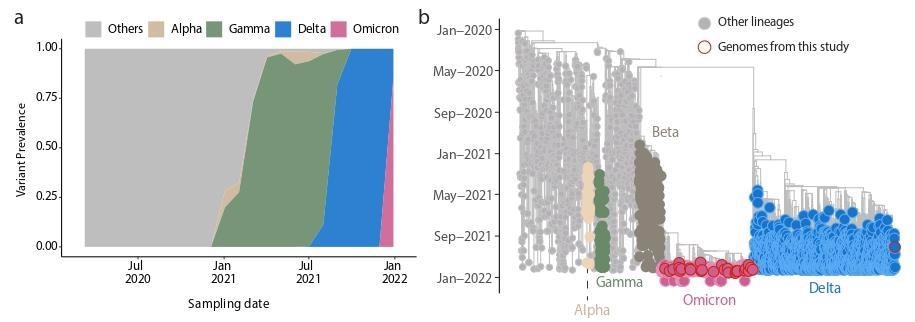
Genomic epidemiology of the SARS‐CoV‐2. Omicron variant in Salvador, Bahia, Northeast Brazil. A) Dynamics of the SARS-CoV-2 epidemic in Bahia showing the progression in the proportion of circulating variants in the state over time, with the rapid replacement of the Delta by the Omicron variant; B) Time-resolved maximum likelihood phylogenetic tree including the newly (n=29) Omicron isolates obtained in this study plus n = 4,249 representative SARS‐CoV‐2 genomes collected up to January 16th, 2021. Alpha, Beta, Gamma, Delta and Omicron VOCs are highlighted in the tree. Newly Omicron and Delta genomes obtained in this study are highlighted with a red border. Genomes of other lineages are shown in grey.
Among the vaccinated participants, the analysis revealed that 99% of cases were due to Omicron and the Delta variant was responsible for just one COVID-19-case. Among unvaccinated individuals, all four participants were Omicron infected.
In Bahia, three VOCs primarily dominated the epidemiology, as revealed by the genomic surveillance analysis. The Gamma variant (P.1) dominated in Bahia until August 2021, before being replaced by Delta (B.1.617.2); finally, Omicron replaced the Delta variant to become the predominant variant in Bahia in December 2021. Although present, the SARS-CoV-2 Alpha variant remained at a frequency of less than 1% in Bahia.
The researchers constructed a phylogenetic tree with these VOCs to map them to those found in other parts of the world; additionally, explore the relationship between the sequenced genomes. The results of the time-sampled phylogeny revealed that the Omicron was independently introduced multiple times over time in Bahia.
Conclusions
To summarize, the study findings suggested that fully vaccinated patients had a favorable clinical prognosis; however, they become SARS-CoV-2 carriers after a BTI within the community. In the case of Bahia, even double or triple vaccinated individuals developed BTIs from Omicron VOC with a high viral load.
The genome-wide sequencing in the state of Bahia reinforced the notion that viral dissemination generally follows national and international patterns of human mobility, facilitating the spread of emerging VOCs within countries and globally. As of 2020, the co-circulation of three different VOCs such as Gamma (P.1), Delta, and Omicron appeared to have dominated the epidemiological history in the state.
The study puts the notion that herd immunity of over 70% prevents the population-level dissemination of SARS-CoV-2 under scrutiny as Brazil is among those countries that have surpassed this figure. Moreover, Brazil has a relatively high COVID-19 vaccination coverage of 74.9% (with two doses of vaccine). Yet the SARS-CoV-2 reinfection rates, post-emergence of Omicron, were substantially high in Bahia.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Genomic monitoring unveils a high prevalence of SARS-CoV 2 Omicron Variant in vaccine breakthrough cases in Bahia, Brazil, Gubio Soares Campos, Marta Giovanetti, Laise de Moraes, Helena S da Hora, Keila Motta Alcantara, Silvia Ines Sardi, medRxiv, 2022.02.16.22271059; doi: https://doi.org/10.1101/2022.02.16.22271059, https://www.medrxiv.org/content/10.1101/2022.02.16.22271059v1
- Peer reviewed and published scientific report.
Campos, Gúbio Soares, Marta Giovanetti, Laíse de Moraes, Helena Souza da Hora, Antônio Carlos de Albuquerque Bandeira, Keila Veronica Oliveira Motta De Alcantara, and Silvia Ines Sardi. 2023. “Genomic Monitoring Unveils a High Prevalence of Severe Acute Respiratory Syndrome Coronavirus 2 Omicron Variant in Vaccine Breakthrough Cases in Bahia, Brazil.” Revista Da Associação Médica Brasileira 69 (March): 257–61. https://doi.org/10.1590/1806-9282.20220955. https://www.scielo.br/j/ramb/a/3Qpwxdcrrq5sDrBCQkW3pKC/?lang=en.