
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Omicron BA.1 and BA.2 variants emerged and spread rapidly across the globe within a few months. The difference between the pathogenicity, infectivity, and vaccine evasion of the co-circulating Omicron BA.1 and BA.2 variants has not been fully understood. This poses a considerable challenge for effective surveillance, rendering information obtained from cell cultures and whole-genome sequencing insufficient to provide fast and reliable data.
Due to the significant impact of Omicron BA.1 and BA.2 variants on public health, there is a high demand for the development of a diagnostic tool to identify these emerging variants rapidly.
Study design
In this study, nasopharyngeal swab samples of patients were collected and ribonucleic acid (RNA) was extracted for clinical testing. Vero-6 cells were infected with the swab samples, and the cultured virus was analyzed through RNA extraction and successive PCR.
Specific mutations in the Omicron BA.1 and BA.2 variants were identified by aligning complete genome sequences of the SARS-CoV-2 Wuhan (A19), Delta, Omicron (BA.1), and Omicron (BA.2) variants. The researchers obtained genome sequences from the Global initiative on sharing all influenza data (GISAID), and the sequences were aligned and analyzed by the Geneious software package. To ensure the uniqueness of each mutation, selected regions were again analyzed through NextStrain website tools. Probes and primers were designed and tested by the Primer 3 program.
RT-qPCR multiplex assays were performed using specific probes and primers sequences used in the study for each reaction. Various cycling conditions for Omicron reactions (varying temperatures, reaction times, and concentrations) were used and fluorescence was examined in the extension step of each cycle.
The qPCR target sequences spanning region was amplified and then transcribed to RNA using an in vitro transcription kit. Finally, the purification and quantification of the obtained in vitro transcribed (IVT) RNA was performed. For each reaction, calibration curves were generated using synthesized standards, and the analytical limit of detection was estimated.
Findings
The researchers analyzed complete genome sequences of the SARS-CoV-2 Wuhan (A19), Delta (B.1.617.2), Omicron (BA.1), and BA.2 strains. They identified and selected four sequences that were specific and identical for both BA.1 and BA.2 variants, sequences that were unique to either BA.1 or BA.2, and sequences that were absent in Wuhan and Delta genomes. The deletion at 31 amino acid position (N31del) in the nucleocapsid (N) gene was common for both Omicron BA.1 and BA.2.
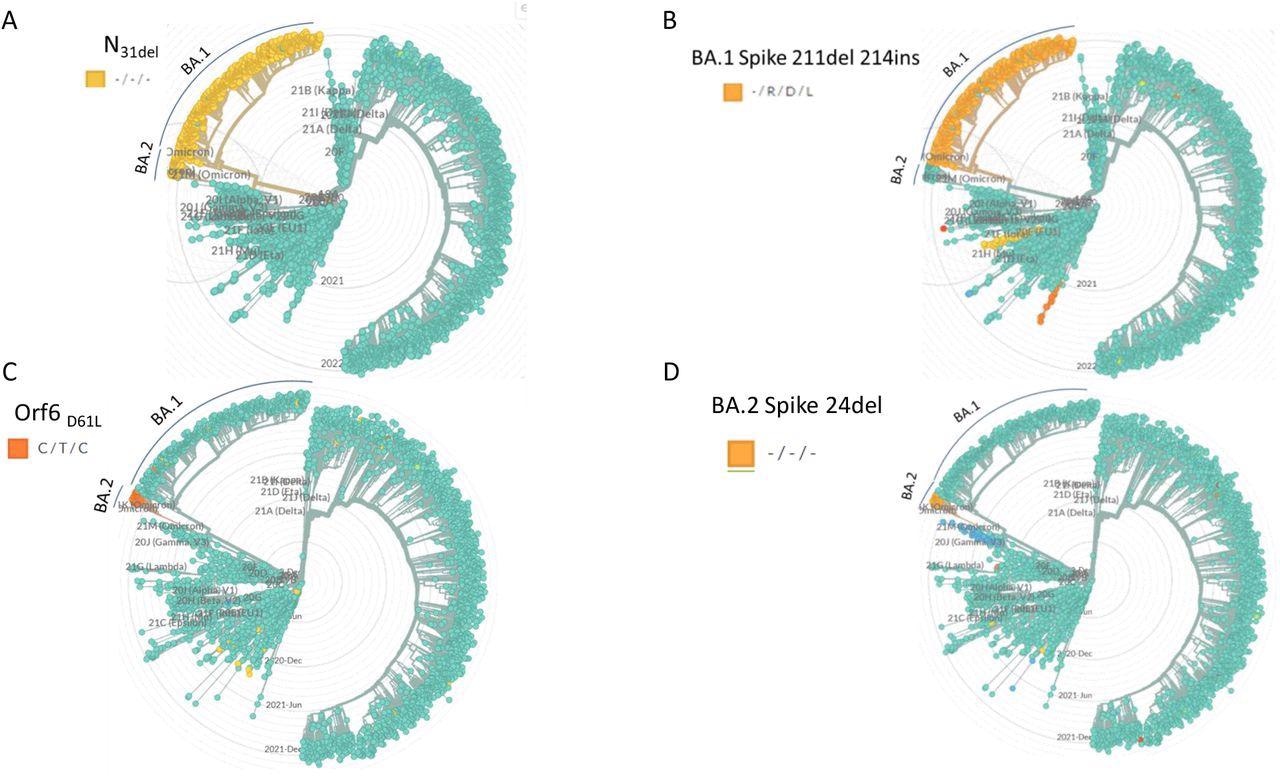
Specificity of selected PCR target mutations. The target mutations selected for qPCR design were identified with respect to circulating strains by using the GISAID-based data with the NextStrain phylogenetic analysis software. The clades that contain the selected mutations are highlighted in each dendrogram. The clade annotation (BA.1, BA.2) is marked with a label and a line.
The deletion at the 211 amino acid position and the insertion at the 214-216 positions in the spike (S) gene were exclusive for the BA.1 variant. While the deletion at the 24 amino acid position in the S gene (S24del) and the codon substation at the 61 amino acid position of the open reading frame (Orf6) gene were both unique to the BA.2 variant. Four probe-based qPCR reactions were designed based on these mutations. The probe in each reaction was complementary to the mutated sequences.
The analytical sensitivity of the new reaction was determined. The S24del reaction was amalgamated in a multiplex containing the E-sarbeco reaction and the human RNAse P reaction. Serial dilution of the IVT targets analogous to the tested multiplex reaction was performed. A total three multiplex reactions were assayed: E-sarbeco + S211del, E-sarbeco + S24del + N31del + hRNAse P, and E-sarbeco + Orf6D61L. Post-analysis of the calibration curve, an analytical limit of detection between one to ten copies per reaction was noted for the hRNAse P, E-sarbeco, S24del, and N31del.
The team observed that out of the 36 clinical samples examined, four samples were characterized as non-BA.1/BA.2 (previously detected as Delta), 10 were recognized as BA.1 (formerly detected as Omicron), and 21 were characterized as BA.2. One sample was positive in the N31del reaction only with a 38.99 quantification cycle (Cq) value and hence categorized as an “unclassified sample”. For the cultured sample and weak clinical sample, the Cq values ranged between nine and 41, respectively. This demonstrated that for clinical samples, the reaction sensitivity was coherent with the results of analytical sensitivity calibration.
Conclusion
The novel RT-qPCR assays developed in this study are sensitive and straightforward in the detection and differentiation of the co-circulating SARS-CoV-2 Omicron BA.1 and BA.2 variants. These assays may be utilized in research and diagnostic laboratories as a tool for testing environmental and clinical samples
The new assays have several advantages which include specific reactions and the ability to distinguish between BA.1/BA.2 and non-BA.1/BA.2 in a single assay. Moreover, the assays were able to differentiate between closely related but not identical variants. Lastly, they detected BA.1 and/or BA.2 RNA while testing environmental samples that represented a heterogeneous pool of various individual sources.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
NOVEL RT-qPCR ASSAYS ENABLE RAPID DETECTION AND DIFFERENTIATION BETWEEN SARS-COV-2 OMICRON (BA.1) AND BA.2 VARIANTS, Oran Erster, Areej Kabat, Hadar Asraf, Virginia Levy, Batya Mannasse, Roberto Azar, Ital Nemet, Limor Kliker, Shay Fleishon, Michal Mandelboim, Ella Mendelson, Neta S Zuckerman, medRxiv, 2022.02.22.22271222; doi: https://doi.org/10.1101/2022.02.22.22271222, https://www.medrxiv.org/content/10.1101/2022.02.22.22271222v1
- Peer reviewed and published scientific report.
Erster, Oran, Ella Mendelson, Virginia Levy, Areej Kabat, Batya Mannasse, Hadar Asraf, Roberto Azar, et al. 2021. “Rapid and High-Throughput Reverse Transcriptase Quantitative PCR (RT-QPCR) Assay for Identification and Differentiation between SARS-CoV-2 Variants B.1.1.7 and B.1.351.” Edited by Tulip Jhaveri. Microbiology Spectrum 9 (2). https://doi.org/10.1128/spectrum.00506-21. https://journals.asm.org/doi/10.1128/Spectrum.00506-21.