The ongoing COVID-19 pandemic entered a new phase when concerns regarding the prolonged efficacy of SARS-CoV-2 vaccines emerged. This inference was associated with the increased incidences of breakthrough infections by heavily mutated SARS-CoV-2 variants in highly vaccinated populations and uncertainty about long-term vaccine effectiveness. Hence, novel treatment targets are a significant resource in the fight to minimize the mortality and morbidity linked to the SARS-CoV-2-induced COVID-19 pandemic.
Most of the currently used COVID-19 treatment strategies work either by boosting host immune responses, reducing viral replication, or decreasing hyperinflammation. However, only a few approaches have attempted to alter a virus-interacting host protein. Further, although genome-wide association studies (GWAS) discovered risk loci, some loci are linked to co-morbidities and are not selective to host-virus interactions.
Study: Low expression of EXOSC2 protects against clinical COVID-19 and impedes SARS-CoV-2 replication. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
In the current study, the scientists identified and experimentally confirmed an association between the reduction in the expression of EXOSC2 in humans and lowered SARS-CoV-2 replication. They conducted an unbiased genetic screening of 332 previously identified genes encoding host proteins interacting with SARS-CoV-2 proteins. For this, the team employed genotype-tissue expression (GTEx)-derived expression quantitative trait loci (eQTL) data.
The association between reduced EXOSC2 expression and inhibition of SARS-CoV-2 replication was confirmed using genetically engineered Calu-3 cells of the EXOSC2. Further, transcriptome analysis was also conducted.
Findings
The results showed that all the 332 host proteins evaluated, including EXOSC2, directly interacted with the SARS-CoV-2 proteins. For 208 of the 332 host proteins, lung-specific eQTLs were discovered using GTEx (v7). Combining COVID-19 GWAS results for gene-specific eQTLs established a link between higher EXOSC2 expression and an increase in the probability of clinical COVID-19 that passed various testing corrections. EXOSC2 interacted with the non-structural protein 8 (Nsp8) present in the SARS-CoV-2 ribonucleic acid (RNA) polymerase.
EXOSC2 was a human RNA exosome component. The SARS-CoV-2 RNA polymerase interacted with most of the human RNA exosome constituents, according to the liquid chromatography with tandem mass spectrometry (LC-MS-MS) protein pulldown analysis. The host RNA exosome components that interacted with SARS-CoV-2 RNA polymerase in Strep-tagged Nsp8 pulldowns were EXOSC10, EXOSC9, EXOSC8, EXOSC7, EXOSC6, EXOSC5, EXOSC4, EXOSC3, EXOSC2, and EXOSC1. Further, similar to EXOSC2, an increase in the expression of EXOSC9 and EXOSC7 was also linked to a high chance for clinical COVID-19.
Nonsense mutations in Calu-3 cells within EXOSC2 were introduced using CRISPR/Cas9 and resulted in a reduction in EXOSC2 protein production by the cells. This reaction hampered the replication of SARS-CoV-2 and increased oligoadenylate synthase (OAS) genes, all of which resulted in an effective immune response targeting SARS-CoV-2. However, host cell viability was not compromised by reduced expression of EXOSC2. Changes in OAS gene expression occurred in the absence of considerable elevation of other interferon-stimulated genes (ISGs) and autonomous to SARS-CoV-2 infection.
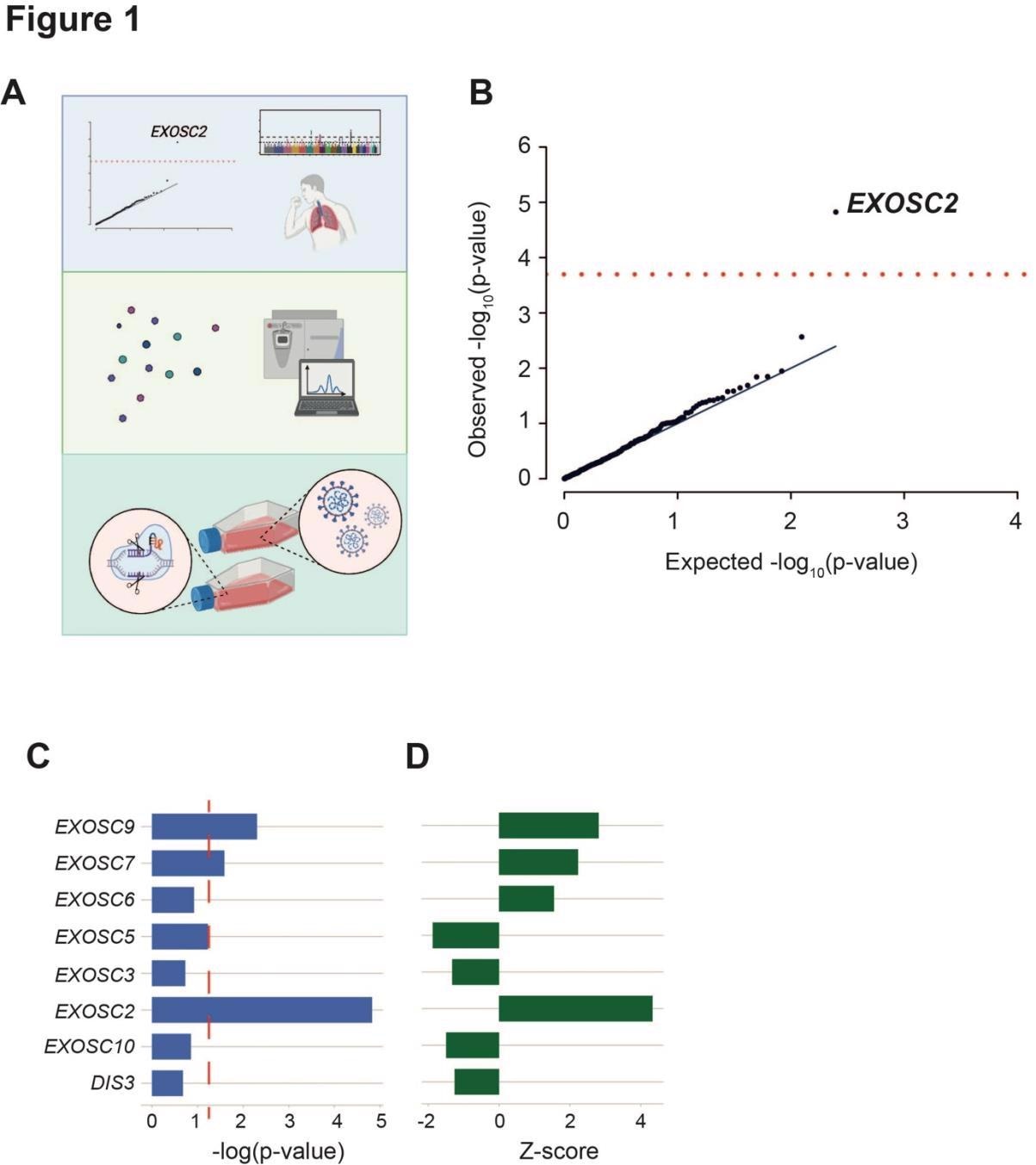
An unbiased screen of host proteins identified as high confidence interacting partners of SARS-CoV-2 proteins links RNA exosome components to the risk of clinical COVID-19. (a) Schematic of the study design. Known host-viral interactions were screened for disease-association by combining lung-specific eQTLs with a GWAS for COVID-19 symptoms. Identification of a positive correlation between EXOSC2 expression and increased severity of COVID-19 led to further study of interactions between the SARS-CoV-2 polymerase and the entire human RNA exosome by AP-MS. Finally, CRISPR editing of EXOSC2 within human lung cells and subsequent infection with SARS-CoV-2 facilitated validation of the relationship between EXOSC2 expression and viral replication and interrogation of the underlying biological mechanism. (b) Lung eQTLs were used to group genetic variants according to their effect on the expression of 332 host genes encoding proteins that interact with viral proteins. Only expression of EXOSC2 was significantly associated with clinical risk of COVID-19 after Bonferroni multiple testing (red line). (c-d) Lung eQTLs were used to group genetic variants according to their effect on expression of all genes encoding components of the RNA exosome. Expression levels of EXOSC7, EXOSC9 and EXOSC2 were significantly linked to clinical COVID-19 and in each case, higher expression was associated with a higher risk of infection. p=0.05 is indicated by a red dashed line.
Conclusions
The study findings demonstrate that reduced expression of EXOSC2 was well tolerated in a considerable proportion of the population who were comparatively immune to clinical COVID-19. Moreover, excess toxicity was not observed in genetically modified Calu-3 cells. However, the authors failed to identify the mechanism relating to the interaction of SARS-CoV-2 with the host RNA exosome and alterations in viral replication. Hence, further structural biology investigations are required.
Nonetheless, the authors proposed several potential mechanisms involved in SARS-CoV-2-human RNA exosome interaction and changes in SARS-CoV-2 replication. The present findings suggested that reduced SARS-CoV-2 replication in EXOSC2-depleted cells was not significantly linked to increased baseline ISG expression in these cells. This is because the OAS gene was upregulated even in the absence of ISGs.
Although the study did not evaluate whether the alterations in the expression of OAS protein modulate the impact of EXOSC2 on SARS-CoV-2 replication, it could be the mechanism associating SARS-CoV-2 replication and EXOSC2 expression.
To summarize, the study detected a novel therapeutic target, EXOSC2, against SARS-CoV-2 infection. It indicated that targeted EXOSC2 inhibition or depletion might be an effective and safe way to protect at-risk populations from clinically symptomatic COVID-19. The current research might add new insights to COVID-19 therapy and influence the development of novel treatment strategies against SARS-CoV-2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Low expression of EXOSC2 protects against clinical COVID-19 and impedes SARS-CoV-2 replication, Tobias Moll, Valerie Odon, Calum Harvey, Mark Collins, Andrew Peden, John Franklin, Emily Graves, Jack Marshall, Cleide dos Santos Souza, Sai Zhang, Mimoun Azzouz, David Gordon, Nevan Krogan, Laura Ferraiuolo, Michael Snyder, Pamela Shaw, Jan Rehwinkel, Johnathan Cooper-Knock, bioRxiv, 2022.03.06.483172; doi: https://doi.org/10.1101/2022.03.06.483172, https://www.biorxiv.org/content/10.1101/2022.03.06.483172v1
- Peer reviewed and published scientific report.
Moll, Tobias, Valerie Odon, Calum Harvey, Mark O. Collins, Andrew Peden, John Franklin, Emily Graves, et al. 2023. “Low Expression of EXOSC2 Protects against Clinical COVID-19 and Impedes SARS-CoV-2 Replication.” Life Science Alliance 6 (1). https://doi.org/10.26508/lsa.202201449. https://www.life-science-alliance.org/content/6/1/e202201449.