As of March 21st, 2022, the causative virus of coronavirus disease 2019 (COVID-19), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has infected over 470 million people worldwide and claimed the lives of over 6 million. Most persons infected with SARS-CoV-2 are asymptomatic or show mild flu-like symptoms. In a prospective study of adults with SARS-CoV-2 infection, 91% were asymptomatic or had mild disease as outpatients, while only 9% required inpatient care.
COVID-19 immunopathology is characterized by lymphopenia, lymphocyte dysfunction, innate immune cell abnormalities, and elevated cytokine production. Early investigations of serum cytokine levels in COVID-19 patients showed increased amounts of circulating Interleukin 6 (IL-6), leading to the idea of an IL-6-driven cytokine storm and the immunopathology that resulted. Clinical trials using IL-6 neutralizing therapies show significant clinical benefits, with additional benefits in patients receiving supplemental corticosteroid treatment, implying that immune signaling and immune cell activation modulation have clinical significance for disease intensification and resolution.
In a recent study published on the bioRxiv* preprint server, a group of researchers from the University of California, San Francisco, looked at intra-patient immunological changes over time to see if there were any changes in immune responses associated with successful COVID-19 resolution. The researchers took longitudinal peripheral blood samples from COVID-19 patients in hospitals, SARS-CoV-2 negative ventilated patients, and healthy people. The authors used mass cytometry and a unique panel of antibodies specific for immune cell phenotyping and detecting phosphorylated cell signaling proteins to explore changes in immune cell signaling states across time.
 Study: A conserved immune trajectory of recovery in hospitalized COVID-19 patients. Image Credit: NIAID
Study: A conserved immune trajectory of recovery in hospitalized COVID-19 patients. Image Credit: NIAID

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Longitudinal peripheral blood sampling from hospitalized patients both positive and negative for COVID-19
This study gathered longitudinal peripheral blood (PB) specimens from patients with COVID-19 and patients who were negative for COVID-19 who were admitted to UCSF Medical Center and Zuckerberg San Francisco General Hospital to investigate the proportion of circulating immune cells and cell signaling states that characterize SARS-COV-2 infections and distinguish them from other respiratory infections. Patient demographics and clinical factors are linked to PB samples. As controls, healthy people's PB samples were acquired. To measure the level of 30 protein markers and 14 phosphorylated signaling molecules, all samples were processed, stained, and analyzed by mass cytometry.
The final cohort of 230 samples consisted of 205 samples from 81 COVID-19 patients, 14 samples from 7 COVID-19 negative patients, and single samples from each of 11 healthy patients who satisfied the quality control standards. Patients with COVID-19 were divided into COVID-19 severity groups depending on their WHO score on the day of sampling. The researchers manually gated 38 canonical immune cell groups based on the phenotypic indicators in the antibody panel and examined immune cell population frequencies, protein expression patterns, and immune cell signaling pathways relevant to COVID-19 course progression and resolution.
The innate immune arm of COVID-19 infection shows distinct compartmental alterations in monocyte and neutrophil composition
Because there were significant differences in innate immune compartments between patients with COVID-19, patients with other respiratory illnesses, and healthy controls, the investigators looked into the makeup of neutrophils and monocytes. While the number of neutrophils in COVID-19 patients and healthy people did not differ considerably, the authors discovered that the expression of a number of proteins on neutrophils differed between the two groups.
The expression of CD11c, CD14, CD16, and PD-L1 in COVID-19 patients' neutrophils was considerably higher, indicating a highly activated and inflammatory neutrophil phenotype. Furthermore, while the frequency of all monocytes was similar between groups, the makeup of monocyte subsets was significantly different between COVID-19 and other respiratory infections patients and healthy people. The frequency of intermediate monocytes increased substantially in patients, while the frequency of classical monocytes decreased.
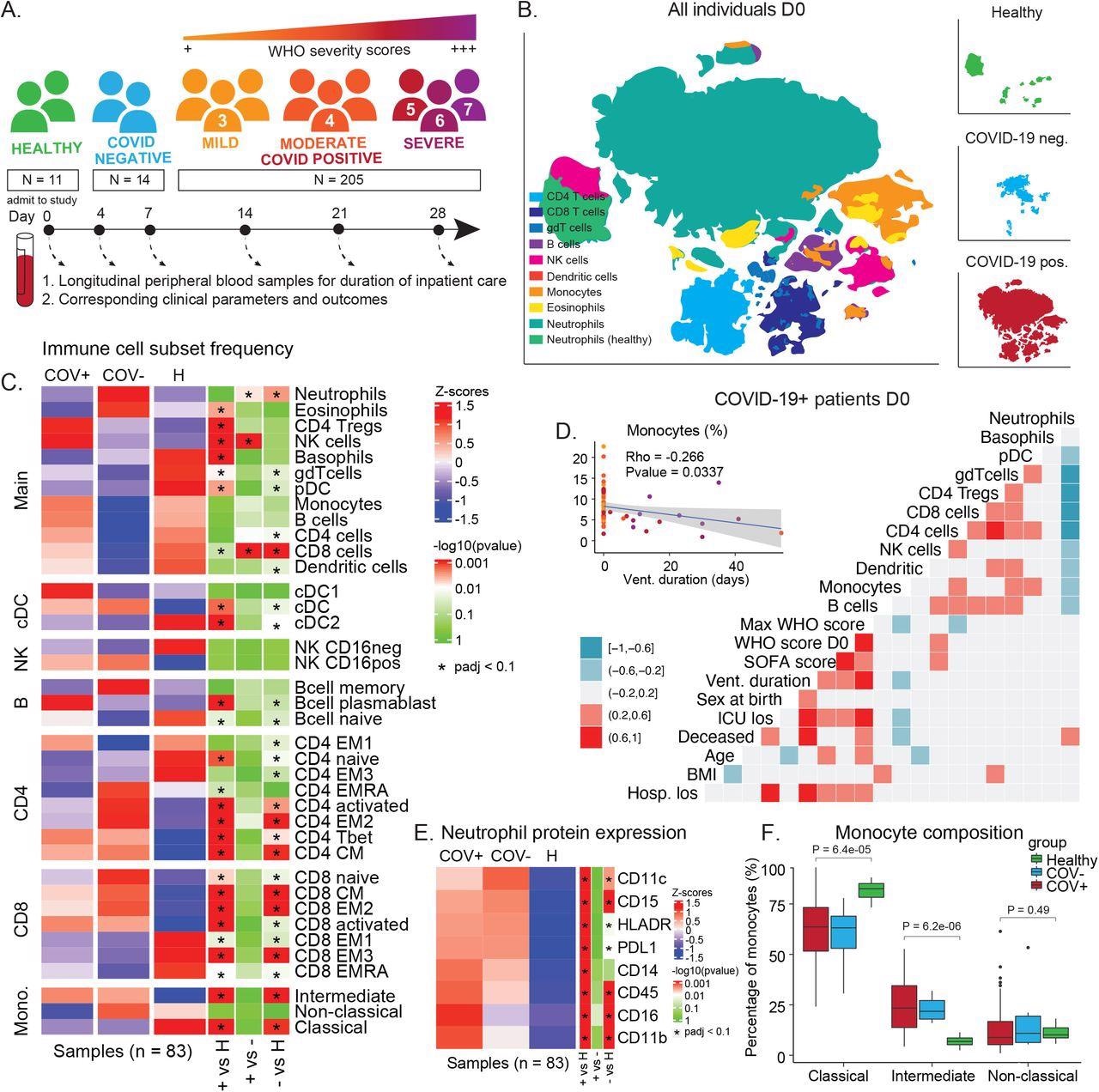
COVID-19 immune phenotype and composition is highly divergent from healthy individuals and has unique features compared to other severe respiratory infections. A) Overview of cohort. Patients were admitted to the hospital and enrolled in the study at D0. Peripheral blood samples were collected throughout the duration of stay. Corresponding clinical parameters and WHO scores were documented. 205 samples from 81 COVID-19 positive patients were included in the final cohort. Additionally, 14 samples from 7 COVID-19 negative patients with other respiratory diseases and 11 healthy individuals were included in the study. B) t-SNE plot of all patient samples at D0 (n = 83) using phenotypic markers colored by major immune cell populations. Upper right panel: t-SNE plot of healthy samples (n = 11); middle right panel: t-SNE plot of COVID-19 negative samples (n = 6); lower right panel: t-SNE plot of COVID-19 positive samples (n = 66). C) Immune cell population abundance at D0 in COVID-19 positive (+), COVID-19 negative (-) patients, and healthy individuals (H). P-values obtained by Wilcoxon Rank Sum Test, followed by Benjamini-Hochberg correction with FDR < 0.1. D) Correlation between cell population abundance at D0 and clinical outcomes, e.g. ventilation duration (vent_duration) and hospital length of stay (hosp_los) for COVID-19+ patients. Correlation estimates are obtained by Pearson correlation. E) Protein expression on neutrophils (F) in COVID-19 positive (COV+), COVID-19 negative (COV-) patients, and healthy controls at D0 (Wilcoxon Rank Sum Test, Benjamini-Hochberg correction with FDR < 0.1). F) Frequency of monocyte subsets in COVID-19 positive (COV+), COVID-19 negative (COV-) patients, and healthy controls at D0. P-values obtained by Wilcoxon Rank Sum Test.
COVID-19 resolution is linked to increased cell signaling at the time of admission
To see if observed alterations in cell frequencies were accompanied by defective signaling, the researchers looked at signaling dynamics in late discharge and eventually died individuals. In contrast to patients who resolved COVID-19 in less than 30 days, who showed consistent changes in signaling states from high to low over time, they found no meaningful differences in late discharged and eventually deceased patients. Instead, these individuals had asymmetric signaling directionality in activated CD8 T cells, no pS6 signaling in cDC1 cells, and reduced tp1 signaling across monocyte subsets.
Surprisingly, when late discharged patients are within 30 days of discharge, the trajectory of several immune resolution features, such as monocytes, neutrophils, and signaling molecules, resembles the recovery trajectories in patients hospitalized less than 30 days, implying that the resolution phase begins in these patients as well. These findings suggest that late discharge and eventually dead patients have lower immune cell signaling at the time of admission. While some of these cell signaling pathways in these patients became more active with time, others remained unchanged.
Implications
This study presents a foundational model of a successful anti-SARS-CoV-2 immune response, as well as an understanding of the core immunological alterations that accompany disease recovery after severe COVID-19. This working model of a recovering immune response trajectory can be used to contextualize divergent immunological processes in immunosuppressed or immunocompromised patients with poor disease outcomes, long-haul COVID-19 patients, or response to novel variants. In addition, this study highlights essential immunological pathways that could be addressed to enable recovery of severe disease in COVID-19 patients and possibly other acute respiratory infections by delineating a conserved trajectory of effective recovery.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.