In a recent study posted to the bioRxiv* preprint server, researchers conducted a phylogenetic investigation of the human parainfluenza type 3 virus strain (HPIV3) that caused an outbreak in South Korea amid the coronavirus disease 2019 (COVID-19) pandemic.
 Study: Phylogenetic analysis of human parainfluenza type 3 virus strains responsible for the outbreak during the COVID-19 pandemic in Seoul, South Korea. Image Credit: Experienceplus / Shutterstock
Study: Phylogenetic analysis of human parainfluenza type 3 virus strains responsible for the outbreak during the COVID-19 pandemic in Seoul, South Korea. Image Credit: Experienceplus / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
HPIVs are significant respiratory pathogens that render acute respiratory infections in children and babies. There are four types of HPIVs: HPIV1, HPIV2, HPIV3, and HPIV4. HPIV3, which comes under the Respirovirus genus in the Paramyxoviridae family, is the most frequent kind of HPIVs and is typically linked to lower respiratory tract infections (LRTIs). HPIV3 infections have a distinct seasonal pattern, peaking in late spring and summer. However, in Korea, an out-of-season HPIV3 resurgence has been seen from September 2021. This was despite the presence of the COVID-19-related non-pharmaceutical interventions (NPIs).
About the study
In the present study, scientists looked at the molecular features of the HPIV3 strains that caused an outbreak in Seoul, South Korea, since September 2021. They also investigated the molecular evolution of HPIV3 discovered during the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. The team collected 61 nasopharyngeal swabs (NPS) samples positive for HPIV3 during October and November 2021 from the Korea University Guro Hospital. NPS specimens positive for HPIV3 were identified through a multiplex real-time real transcription-polymerase chain reaction (rt-PCR) assay, called Anyplex II RV16 Detection Kit.
Clinical information, like patient demographics, was gathered from the hospital's clinical records. HPIV3 ribonucleic acid (RNA) was extracted from the NPS specimens using Microlab STARlet, a nucleic acid extraction platform. Further, complementary deoxyribonucleic acid (cDNA) was generated via the RevertAid First Strand cDNA Synthesis kit. The HPIV3 hemagglutinin-neuraminidase (HN) genes' partial nucleotide sequences were matched with previously reported HN gene sequences for genetic distance (p-distance) and phylogenetic evaluations employing 33 HPIV3-positive specimens.
Results
The results indicated that of the 61 HPIV3-positive NPS samples, 29 and 32 belonged to female and male patients, respectively. The median age of the subjects was two years. LRTI symptoms, like bronchiolitis and pneumonia, were seen in 6.6% and 34.4% of patients, respectively. In children and infants, the most common diagnosis was upper respiratory tract infection (URTI), which was succeeded by croup, bronchiolitis, and pneumonia. The HPIV3's prevalence pattern in 2021 in South Korea was distinct from the last five years, with an unanticipated surge starting in September, lasting throughout the winter, and a high positivity rate.
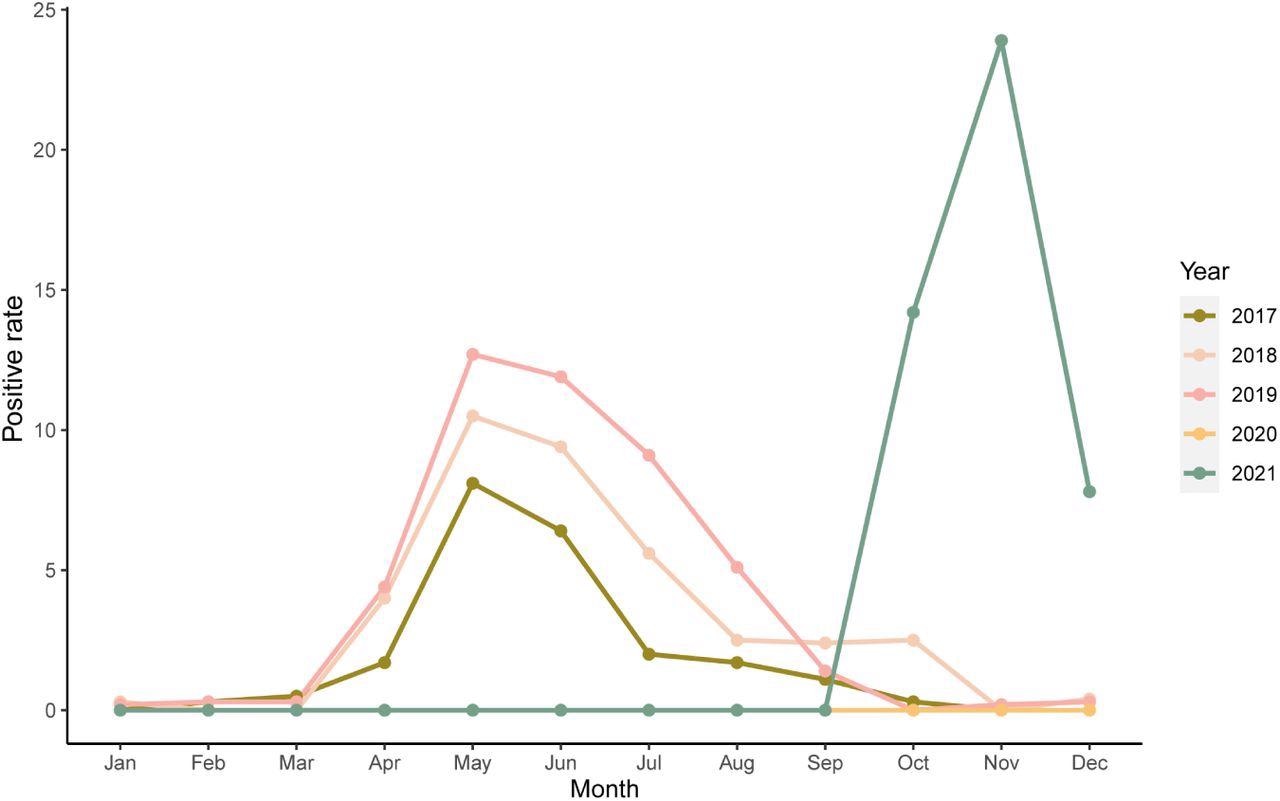
Seasonality pattern of human parainfluenza virus 3 (HPIV3)-positive cases per month between 2017 and 2021. The positivity rate was defined as the number of HPIV3 positive cases compared to the total number of multiplex real-time polymerase chain reaction tests.
According to the phylogenetic tree, all Seoul HPIV3 strains belong to the phylogenetic subcluster C3. Nevertheless, these strains established a distinct cluster from the C3a lineage, which was named in the study as C3h lineage. With a p-distance of less than 0.001, this cluster had 99% bootstrap support. The genetic distances varied from 0.013 in C3a to 0.023 in C3c in other C3 lineages.
As per the deduced HN gene's amino acid sequences, the four protein alterations in Seoul HPIV3 strains (E514K, G387S, K31N, and A22T) were seldom seen in other reference strains. Only the KJ672532, C3d reference strains had the E514K mutation, which was found in four Seoul HPIV3 strains. In four Seoul HPIV3 strains (HPIV3/SEL29, 26, 22, and 21), G1540R, a heterozygous mutation was found at the equivalent nucleotide position of 1540. The forward and reverse amplicon reads confirmed this heterozygosity. Finally, 257 and 15 negative and positive selection sites were predicted, respectively, using fast unconstrained Bayesian approximation (FUBAR) and mixed-effects model of evolution (MEME) methods. The predicted negative selection site included G387S and A22T substitutions.
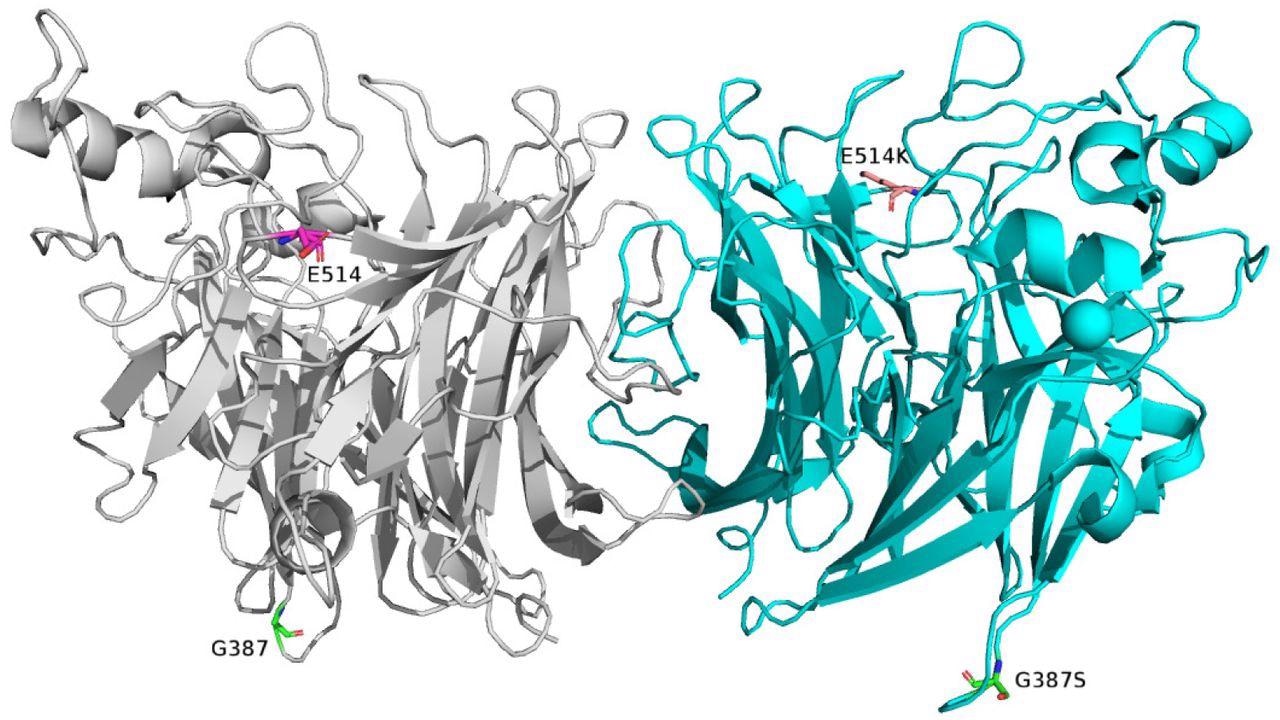
Molecular structure of the HPIV3 HN protein with monomers colored gray (left) and cyan (right). The structure was visualized using PyMol v2.0 (PDB no. 1V3B). The locations of G387 and E514 are highlighted as sticks in the left monomer, and G387S and E514K mutations after mutagenesis are indicated in the right monomer.
Conclusions
The study findings indicated that the HPIV3 peak positivity rate elevated about two-fold relative to the pre-SARS-CoV-2 pandemic phase during 2021 in Seoul. Furthermore, the phylogenetic investigations revealed that the Seoul HPIV3 strains belonged to a different cluster inside subcluster C3. Within subcluster C3, genetic gaps between the strains indicated the emergence of a novel genetic lineage. The appearance of a new genetic lineage might signal the start of a new pandemic.
As a result, due to the resurgence of respiratory pathogens, it is vital to plan for unanticipated viral epidemics and magnitudes. Although the implications of the emergence of this unique HPIV3 lineage are unknown, the current work provides deep insights into the alterations in HPIV3 that occur during the unseasonal epidemics. In addition, further surveillance of circulating HPIV3 strains is required to understand the significance of newly identified mutations.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Phylogenetic analysis of human parainfluenza type 3 virus strains responsible for the outbreak during the COVID-19 pandemic in Seoul, South Korea, Ha Nui Kim, Soo-Young Yoon, Chae Seung Lim, Chang Kyu Lee, Jung Yoon, bioRxiv 2022.03.15.484550, DOI:https://doi.org/10.1101/2022.03.15.484550, https://www.biorxiv.org/content/10.1101/2022.03.15.484550v1
- Peer reviewed and published scientific report.
Kim, Ha Nui, Soo-Young Yoon, Chae Seung Lim, Chang Kyu Lee, and Jung Yoon. 2022. “Phylogenetic Analysis of Human Parainfluenza Type 3 Virus Strains Responsible for the Outbreak during the COVID-19 Pandemic in Seoul, South Korea.” Journal of Clinical Virology 153 (August): 105213. https://doi.org/10.1016/j.jcv.2022.105213. https://www.sciencedirect.com/science/article/pii/S1386653222001469.