COVID-19 respiratory infection causes mild or asymptomatic disease in most individuals. However, moderate to severe disease is observed in nearly 15% of cases, while critical illness is seen in 5% of total cases. During the acute infection phase, myeloid cells like the monocytes/macrophages are extensively enriched in the lungs of COVID-19 patients.
Monocytes are phagocytic, circulating, innate immune cells involved in pathogen sensing and activating innate and adaptive immunity in response to viral infections. Monocytes differentiate into macrophages and dendritic cells (DCs) in affected tissues and contribute to pathogen clearance and tissue regeneration. Several studies reported dysregulated innate responses against SARS-CoV-2, with the myeloid cells exhibiting impaired expression of human leukocyte antigen (HLA)-DR isotype.
 Study: Transcriptional reprogramming from innate immune functions to a pro-thrombotic signature upon SARS-CoV-2 sensing by monocytes in COVID-19. Image Credit: MohamadOuaidat / Shutterstock
Study: Transcriptional reprogramming from innate immune functions to a pro-thrombotic signature upon SARS-CoV-2 sensing by monocytes in COVID-19. Image Credit: MohamadOuaidat / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The study and findings
In this study, researchers evaluated the phenotypic and functional features of classical (CD14+) monocytes in COVID-19 patients relative to healthy controls in the present study. High dimensional flow cytometry evaluated major histocompatibility complex (MHC) molecules and costimulatory and coinhibitory receptors. Although some global phenotypes overlapped across the three groups, monocytes from healthy controls were distinct from mild and moderate COVID-19 patients.
Monocytes from moderate COVID-19 samples demonstrated reduced expression of HLA-DR, but HLA-ABC expression was elevated relative to mild COVID-19 and control samples. Monocytes from moderate COVID-19 samples exhibited reduced expression of CD86, a costimulatory receptor, with an increase in inhibitory receptors – T-cell immunoglobulin and mucin-domain containing-3 (TIM-3) and programmed death-1 (PD-1).
The researchers observed 16 different monocyte subpopulations (clusters). The distribution of cells across the three groups differed in each cluster, revealing distinct monocyte populations in mild and moderate COVID-19 patients. The gene expression activation or repression patterns were explored by studying the epigenetic marks linked with active transcription such as H3K27Ac (histone acetylation) and H3K4Me3 (histone methylation) and gene repression such as H3K9Me2 and H3K27Me3.
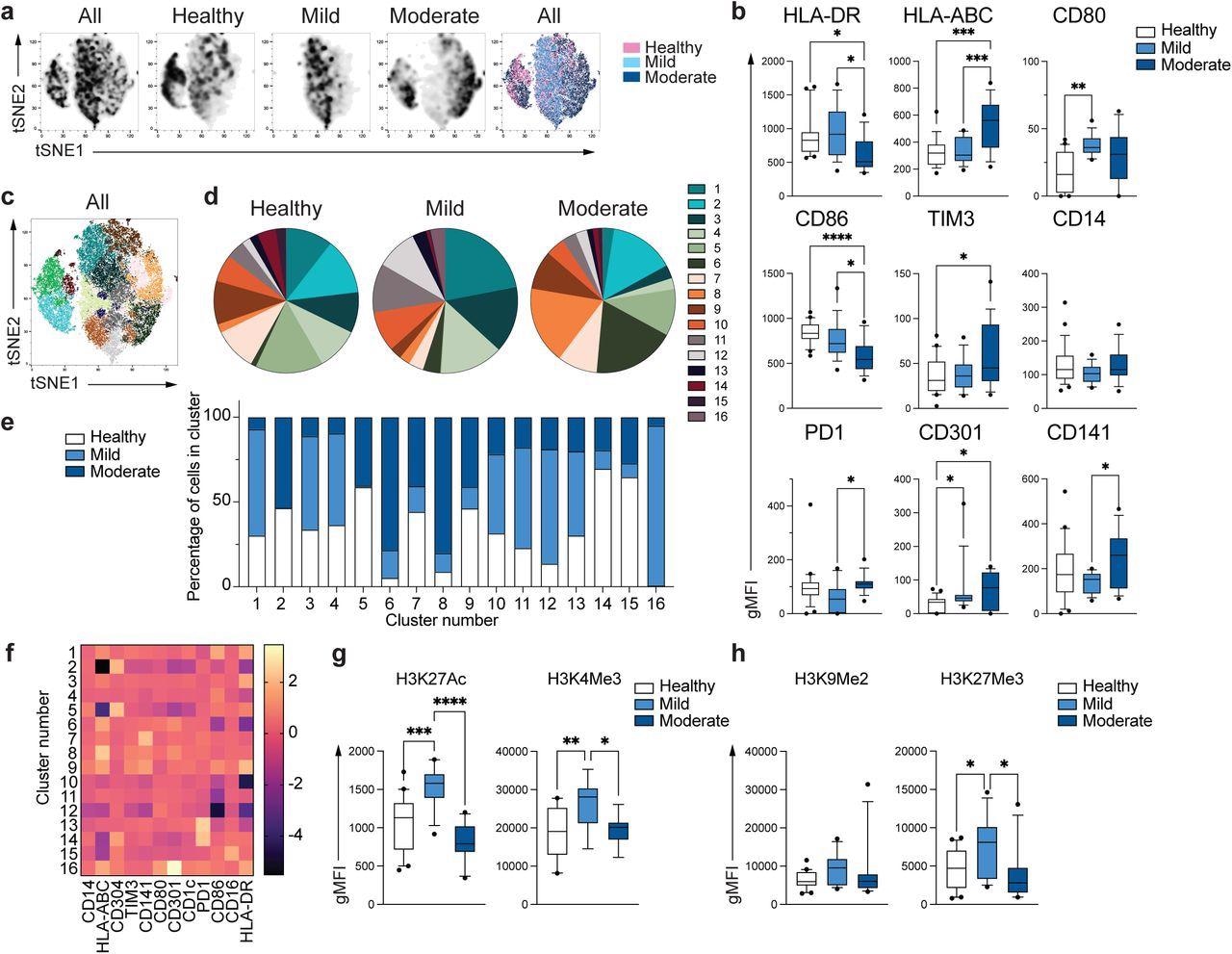
Unique phenotype of COVID-19 monocytes. a. tSNE plots obtained from a concatenated sample consisting of PBMC from n=15 healthy individuals, n=15 mild and n=15 moderate COVID-19 patients. b. Box and whiskers plots summarizing the median gMFI of the receptors analyzed. The box extends from the 25th to the 75th percentile and the whiskers are drawn down to the 10th percentile and up to the 90th percentile. Points below and above the whiskers are drawn as individual points (n=25 healthy, n=15 mild and n=17 moderate COVID-19 individuals). c. tSNE plots depicting the cell clusters identified by Phenograph from the concatenated sample in a. d. Pie charts show the fraction of cells within each identified cell cluster in each patient group. e. Bars graph show the distribution (percentage) of cells from each patient group in each identified cell cluster. f. Heatmap of the expression of receptors per cell cluster displayed as modified z-scores using median values. g and h. Summary of expression of activating (g) and repressive (h) histone marks in monocytes from healthy individuals (n=20), mild (n=15) and moderate (n=11) COVID-19 patients. One-way ANOVA with Tukey’s correction for multiple comparisons for b, g, h. *P<0.05, **p<0.005, ***p<0.001, ****p<0.0001.
Mild COVID-19 monocytes exhibited increased H3K27Ac and H3K4Me3 levels compared to controls, but moderate monocytes displayed expression comparable to healthy controls. There was no difference in the expression of H3K9Me2, a repressive mark; H3K27Me3 expression was increased in mild monocytes compared to controls, not seen in moderate COVID-19 monocytes. These suggested the defective epigenetic remodeling and subsequent activation of innate immune functions in patients with moderate COVID-19.
The gene expression profile of classical monocytes from moderate COVID-19 patients was evaluated in depth relative to healthy controls. Analysis of differential expression of genes revealed the upregulation of 422 genes and downregulation of 187 genes in COVID-19 monocytes relative to healthy subjects. The pathway enrichment analysis (PEA) of upregulated genes (in COVID-19 monocytes) showed a significant increase in lipid metabolism, interferon (IFN), and cytokine signaling.
The increased type I IFN gene signatures in COVID-19 monocytes were confirmed ex vivo by the increased expression of phopho-IFN regulatory factor 3 (IRF3) as well as IFN-induced transmembrane protein 2 (IFITM2), an IFN-stimulated gene (ISG). PEA performed on a set of downregulated genes revealed that glycolysis was the only significantly downregulated pathway in COVID-19 monocytes. The authors observed dysfunctional metabolic profiles besides the reduced activation of nuclear factor kappa-B (NFκB) and intact type I IFN responses in monocytes from moderate COVID-19 patients.
Next, the monocyte functionality to sense and respond to SARS-CoV-2 ex vivo was tested. Monocytes from healthy controls demonstrated a significant increase in tumor necrosis factor (TNF) and interleukin (IL)-10 production upon stimulation with SARS-CoV-2. Contrastingly, COVID-19 monocytes expressed less TNF than control monocytes, while no differences were observed in IL-10 levels. This reduced TNF expression was not specific to SARS-CoV-2, as stimulation with bacterial lipopolysaccharide (LPS) or common cold coronaviruses (CoVs) too had less TNF expression.
Further, ribonucleic acid sequencing (RNA-seq) was performed on SARS-CoV-2-activated monocytes from moderate COVID-19 patients and healthy controls. About 1,437 and 2,073 genes were up and downregulated, respectively, in the COVID-19 monocytes relative to control monocytes. PEA of upregulated genes revealed significant enrichment of pathways involved in hemostasis and coagulation. The downregulated pathways in COVID-19 monocytes were mostly canonical immunological functions such as IFN signaling, activation of T cell receptor signaling in T cells, and innate immune functions with non-lymphoid cells.
Conclusions
The present study assessed monocytes' metabolic, transcriptomic, and functional characteristics and identified several phenotypic and functional changes in monocytes from COVID-19 patients. Ex vivo, COVID-19 monocytes transcriptionally switched to a pro-thrombotic phenotype from a canonical innate immunological function upon sensing the pathogen.
The underlying basis for the observed dysfunctional phenotype (in COVID-19 monocytes) could be epigenetic and metabolic defects. For instance, the defects in histone acetylation might result from the lack of acetyl groups provided by the glycolytic product, acetyl-coenzyme A, whereas glycolysis is significantly downregulated in COVID-19 monocytes. Furthermore, the factors driving monocytic dysfunction need to be investigated. Overall, the study's findings provided a mechanism by which the dysfunction of innate immunity might contribute to the pathology of COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Transcriptional reprogramming from innate immune functions to a pro-thrombotic signature upon SARS-CoV-2 sensing by monocytes in COVID-19, Allison K Maher, Katie L Burnham, Emma Jones, Laury Baillon, Claudia Selck, Nicolas Giang, Rafael J Argüello, Charlotte-Eve Short, Rachael Quinlan, Wendy S Barclay, Nichola Cooper, Graham P Taylor, Emma E Davenport, Margarita Dominguez-Villar, bioRxiv; DOI: https://doi.org/10.1101/2022.04.03.486830, https://www.biorxiv.org/content/10.1101/2022.04.03.486830v1
- Peer reviewed and published scientific report.
Maher, Allison K., Katie L. Burnham, Emma M. Jones, Michelle M. H. Tan, Rocel C. Saputil, Laury Baillon, Claudia Selck, et al. 2022. “Transcriptional Reprogramming from Innate Immune Functions to a Pro-Thrombotic Signature by Monocytes in COVID-19.” Nature Communications 13 (1): 7947. https://doi.org/10.1038/s41467-022-35638-y. https://www.nature.com/articles/s41467-022-35638-y.