The coronavirus disease 2019 (COVID-19) is capable of causing a wide range of clinical manifestations, spanning the spectrum from asymptomatic disease to fatal outcomes. As a result, there has been considerable interest in developing biomarkers that can differentiate severe disease from a mild disease course.
A new study under consideration for the Scientific Reports journal and posted to the Research Square* preprint server explores the possible use of chromatin remodeling in predicting a severe clinical trajectory for COVID-19.
Study: Differential Chromatin Accessibility in Peripheral Blood Mononuclear Cells Underlies COVID-19 Disease Severity Prior To Seroconversion. Image Credit: Rost9 / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Introduction
A growing amount of evidence has indicated the role of a hyper-inflammatory immune response to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the clinical severity of COVID-19. To this end, the extent of chromatin accessibility and gene expression in peripheral blood mononuclear cells (PBMCs), comprising several subtypes of immune cells, are markers of a molecular-level response to SARS-CoV-2.
Mild illness appears to be associated with poor type I and III interferon (IFN) expression, while severe illness is characterized more often by higher levels of pro-inflammatory factors like interleukin-6 (IL-6) and tumor necrosis factor α (TNF-α), as well as a higher number of monocytes and their associated chemokines and cytokines in peripheral blood.
To explore this, regulatory chromatin offers transcription factor motifs that become more accessible as the inflammatory response begins. Thus, these motifs could act as reliable biomarkers for these changes.
In order to analyze epigenetic changes over time, the characteristics of the maturing immune response can be used to reference the point at which the host is in the host immune response. For example, the appearance of anti-SARS-CoV-2 antibodies signals the development of adaptive immunity, shifting from the innate immune response.
This transition to adaptive immunity typically occurs within two weeks of symptom onset and is concurrent with the improvement in clinical symptoms, unless the patient shows signs of progressive disease. The current study explored the possibility that transcription factor motifs could be biomarkers of early changes that signal beneficial or potentially deleterious immune responses in COVID-19.
Study findings
In the current study, the researchers used bulk and single-cell assays including the Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) and ribonucleic acid sequencing (RNA-seq) for chromatin accessible regions. These assays were performed on samples obtained from patients before and after seroconversion to assess the changes within the PBMCs over time.
The study cohort was selected from the ongoing community-based prospective study cohort Molecular Epidemiological Study of Suspected Infection (MESSI). Four groups were formed including pre-pandemic healthy controls, close contacts of patients with SARS-CoV-2 infection but with negative viral and serologic tests, outpatients with mild SARS-CoV-2 infection who tested positive, and those with moderate illness and a positive COVID-19 test result.
Interestingly, symptom severity, using all reported symptoms, showed close similarities between close contacts who remained seronegative for two or more months after exposure; however, this was noticeably different between mild cases/close contacts as compared to those with moderate illness. In fact, individuals with moderate illness most often reported anosmia, ageusia, headache, feeling unwell, and tiredness as compared to reports of coughing with mild COVID and runny nose in close contacts.
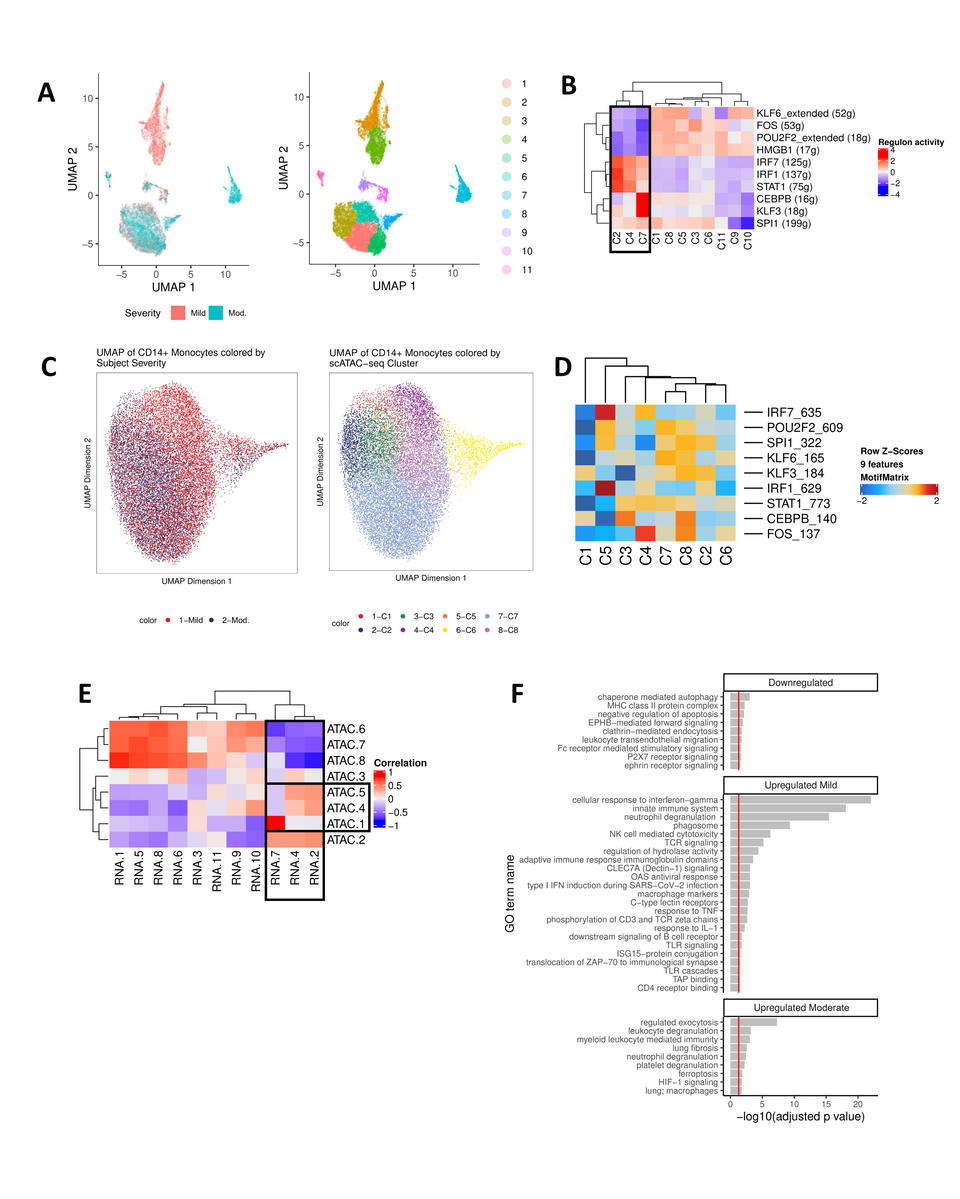
CD14+ monocytes undergo extensive chromatin remodeling prior to seroconversion and harbor severity-specific epigenetic biomarkers. (A) scRNA-seq UMAP of all CD14+ monocytes collected from mild and moderate IgG- subjects. UMAP colored by disease severity (left) and cluster number (right). (B) Heat map depiction of regulon activity computed for each scRNA-seq cluster using SCENIC. Activity of the top 10 regulons (right) is plotted for each cluster of cells from the UMAP (bottom). The black box indicates clusters of interest. (C) scATAC-seq UMAP of all CD14+ monocytes collected from IgG- mild and moderate subjects. UMAP colored by disease severity (left) and cluster number (right). (D) Heat map depiction of transcription factor motif enrichment of identified regulons (right) plotted for each scATAC-seq cluster (bottom). (E) Correlation plot using regulon activity to link clusters between scRNA-seq (columns) and scATAC-seq (rows). Black box indicates clusters of interest. (F) Functional enrichment analysis for regulons down-regulated (KLF6, FOS, POU2F2, HMGB1) or up-regulated (mild: IRF7, IRF1, STAT1; moderate: CEBPB, KLF3) in scRNA-seq clusters 2, 4, and 7 (panel E).
Transcription patterns in PBMCs
The transcription profile in PBMCs differentiated at the gene expression level between cases and healthy controls but not between cases with mild or moderate symptoms. While 30 genes were differentially expressed in seronegative cases with mild or moderate symptoms, 43 differentially expressed genes were found in the seropositive cohort with mild or moderate symptoms.
The IL-8 precursor CXCL8 was expressed at higher levels in controls. When the scientists examined the 455 differentially accessible regions (DARs) in mild and moderate cases who had not yet developed antibodies, they found 73 that showed differential accessibility with the severity of disease, as well as an epigenetic response.
Further analysis showed that mild seronegative cases had a higher level of transcription factors and pathways linked to viral infection as compared to moderate seronegative cases. Both mild and moderate cases showed epigenetic changes that triggered chromatin remodeling without the expected transcriptional activity to follow it. That is, transcription factor motifs were easily accessible, even while gene expression remained low.
Some upregulated genes were identified, most of which regulate inflammation. All PBMC types showed an increase in the accessibility of the domains of regulatory chromatin (DORCs), thus indicating that they were preparing for activation. The cells that showed the highest enrichment of PBMCs in the seronegative period were mostly CD14+ monocytes.
CD14+ monocytes were primed for chromatin remodeling by DORCs regulated by super-enhancers. The regions involved in CD14+ monocytes from those with mild symptoms were enriched for IFN regulatory factor 7 (IRF7), IRF1, and signal transducer and activator of transcription 1 (STAT1) transcription factor regulatory regions as compared with CCAAT enhancer-binding protein beta (CEBPB) and Kruppel like factor 3 (KLF3) regulatory regions in moderate cases.
In seropositive cases, another 375 regions were identified where chromatin showed differential accessibility. Among these, mild cases had more accessible chromatin, thus showing dysregulated myeloid development.
Functional enrichment analysis showed that most transcription factors with differential expression were related to the adaptive immune response, specifically to the shift from innate to acquired immunity. The analysis suggests that B- and T-cell receptors were being activated.
Implications
The results of the current study point to large-scale chromatin remodeling in certain PBMC subsets early in SARS-CoV-2 infection that correspond with mild and moderate symptoms prior to antibody formation. The level of transcription factor activity and chromatin accessibility at this early time point differentiated cases by disease severity, even before actual transcription happened. This occurred mainly in CD14+ monocytes.
The study findings provide more clarity on previous research on chromatin accessibility in severe and recovering COVID-19 cases. The increase in KLF and CREB transcription factor motif accessibility in mild to moderate cases indicates the role of epigenetic priming in monocyte maturation. This has previously been shown to occur in early SARS-CoV-2 infection, while this study shows that their activation level differs with the severity of disease.
Mild disease is associated with rapid viral control and is characterized by the activation of antiviral pathways, such as those mediated by IRF1 and IRF7. Conversely, with more severe disease, dysregulated immune responses begin with ineffective viral control and lower levels of antiviral responses, both of which correspond to moderate disease.
While CD14+ monocytes were the most actively involved in chromatin remodeling over time and with early epigenetic regulation, these motifs were not found to be more accessible after seroconversion during the seronegative period. At this point, antibody-producing cells were more active, thereby indicating the development of adaptive immunity in recovering COVID-19 patients.
“These changes temporally precede transcriptional manifestations of pathways related to adaptive immunity… and suggest that detection of critical components of chromatin remodeling in early disease may offer promise for a new class of diagnostic tools for COVID-19.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Giroux, N. S., Ding, S., McClain, M. T., et al. (2022). Differential Chromatin Accessibility in Peripheral Blood Mononuclear Cells Underlies COVID-19 Disease Severity Prior To Seroconversion. Research Square. doi:10.21203/rs.3.rs-1479864/v1. https://www.researchsquare.com/article/rs-1479864/v1.
- Peer reviewed and published scientific report.
Giroux, Nicholas S., Shengli Ding, Micah T. McClain, Thomas W. Burke, Elizabeth Petzold, Hong A. Chung, Grecia O. Rivera, et al. 2022. “Differential Chromatin Accessibility in Peripheral Blood Mononuclear Cells Underlies COVID-19 Disease Severity prior to Seroconversion.” Scientific Reports 12 (1). https://doi.org/10.1038/s41598-022-15668-8. https://www.nature.com/articles/s41598-022-15668-8.