Various studies have reported several aspects of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific T-cell epitopes. However, extensive research is needed to understand the characteristics of SARS-CoV-2-specific T-cell epitope targets by infection and by vaccine-induced responses.
 Study: SARS-CoV-2-specific T-cell epitope repertoire in convalescent and mRNA-vaccinated individuals. Image Credit: Shutterstock / Design_Cells
Study: SARS-CoV-2-specific T-cell epitope repertoire in convalescent and mRNA-vaccinated individuals. Image Credit: Shutterstock / Design_Cells
About the study
In the present study, researchers estimated SARS-CoV-2-specific T-cell epitope in convalescent individuals who had reported COVID-19 recovery and individuals who had received two and three doses of SARS-CoV-2 Pfizer/ BioNTech messenger ribonucleic acid (mRNA) vaccine.
The team first mapped the overall response of SARS-CoV-2-specific CD8+ T-cell response against human leukocyte antigen (HLA)-restricted SARS-CoV-2-specific CD8+ T-cell epitopes in epitope-specific T-cell cultures. Interferon-γ (IFN-γ) staining was subsequently performed on the cultures. The team also compared the viral sequences in the SARS-CoV-2 wild-type (WT) B and Omicron (B.1.1.529) variants.
To assess the extent of CD8+ T-cell responses among convalescent versus vaccinated individuals, the team examined the overlapping peptides (OLPs) present across the whole SARS-CoV-2 spike protein. For all the positive CD8+ T-cell responses, the researchers estimated the OLP for the HLA-restricted optimal epitopes expressed by each participant. The team also conducted an in silico analysis that predicted the most likely HLA restriction and optimal epitope. CD4+ T-cell responses were also analyzed using OLPs that spanned the S protein.
The team subsequently assessed the impact of booster vaccine- or infection-induced T-cell responses on the S-specific CD8+ T-cell responses by using the overlapping S peptides that mapped S-specific CD4+ and CD8+ T-cell responses in triple-vaccinated as well as convalescent individuals who had received an mRNA booster dose. The observed S-specific CD8+ T-cell responses post mRNA vaccination was analyzed using T-cell responses that targeted highly conserved regions in SARS-CoV-2 among both vaccinated and convalescent persons.
Results
The study results showed that CD8+ T-cell responses among convalescent participants targeted most of the SARS-CoV-2-specific epitopes present in the viral protein. Notably, in vaccinated individuals, the CD8+ T-cell responses were directed against SARS-CoV-2 spike (S) epitopes. Also, a few of the CD8+ T-cell responses targeted non-S epitopes, while the HLA-B*07/N epitope was the main target.
Individual S-specific CD8+ T-cell responses were more likely to be targeted among vaccinated individuals as compared to that in convalescent persons. Moreover, S-specific CD8+ T-cell responses were broader in vaccinated persons than in convalescent ones. On comparing the viral sequences present in SARS-CoV-2 WT and Omicron strains, the team observed that only one optimal CD8+ T-cell epitope was impacted by the variation in viral sequences in Omicron BA.1 and BA.2 sub-variants.
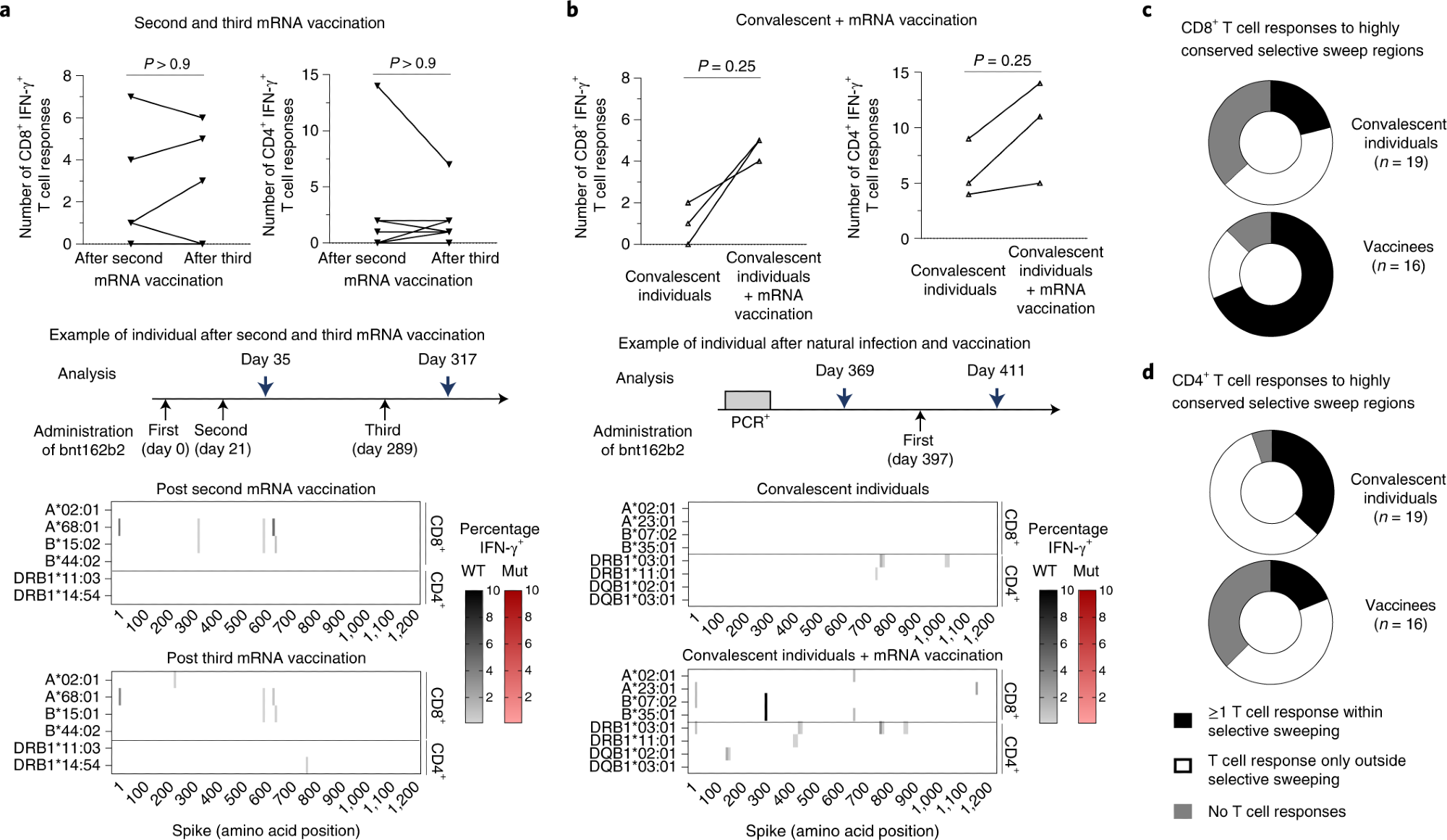 a,b, Number, location and percentages of spike-specific CD8+ and CD4+ T-cell responses to OLPs that are detectable in SARS-CoV-2 vaccinees after the second versus after the third dose of Pfizer/BioNTech mRNA vaccine (bnt162b2, measured 2–4 weeks after vaccination) (a) and in SARS-CoV-2 convalescent individuals who subsequently received a single dose of Pfizer/BioNTech mRNA boost vaccination (bnt162b2, measured 2 weeks after vaccination) (b). The heatmaps depict the data of one representative individual each. Targeted epitopes with sequence variations in Omicron/B.1.1.529, BA.1 are marked in red. c,d, Vaccinees and convalescent individuals with CD8+ (c) and CD4+ (d) T-cell responses within and outside highly conserved selective sweep regions in the spike protein are shown. Statistical analysis was performed with a two-sided Wilcoxon matched-pairs signed-rank test.
a,b, Number, location and percentages of spike-specific CD8+ and CD4+ T-cell responses to OLPs that are detectable in SARS-CoV-2 vaccinees after the second versus after the third dose of Pfizer/BioNTech mRNA vaccine (bnt162b2, measured 2–4 weeks after vaccination) (a) and in SARS-CoV-2 convalescent individuals who subsequently received a single dose of Pfizer/BioNTech mRNA boost vaccination (bnt162b2, measured 2 weeks after vaccination) (b). The heatmaps depict the data of one representative individual each. Targeted epitopes with sequence variations in Omicron/B.1.1.529, BA.1 are marked in red. c,d, Vaccinees and convalescent individuals with CD8+ (c) and CD4+ (d) T-cell responses within and outside highly conserved selective sweep regions in the spike protein are shown. Statistical analysis was performed with a two-sided Wilcoxon matched-pairs signed-rank test.
In convalescent persons, none of the HLA class I alleles showed the restriction of more than two S-specific CD8+ T-cell responses, while in the vaccinated persons, several HLA alleles restricted five or more S-specific CD8+ T-cell responses. Additionally, more S-specific CD8+ T-cell responses were observed per vaccinated person as compared to among convalescent individuals. This indicated the greater extent of CD8+ T-cell responses in the vaccinated cohort.
The team also observed that S-specific CD4+ T-cell responses were more limited after vaccination than after SARS-CoV-2 infection. In particular, a fewer number of S-specific CD4+ T-cell responses were restricted by HLA alleles, while fewer CD4+ T-cell responses were detected in vaccinated individuals as compared to the convalescent persons.
The CD4+ and CD8+ S-specific T-cell responses were comparatively more stable over time in the vaccinated as well as the convalescent persons. Notably, few S-specific CD4+ T-cell responses observed in the vaccinated individuals displayed high conservation between the SARS-CoV-2 WT and Omicron BA.1 and BA.2 variants. Moreover, mRNA vaccination also increased the CD8+ T-cell responses that targeted conserved S-specific epitopes.
The team observed that the selected sweep regions in SARS-CoV-2 had a high extent of amino acid homology among the emerging SARS-CoV-2 variants of concern. Complete homology was found in the SARS-CoV-2 Delta variant, one point mutation was found in the Alpha or Beta variants, two mutations in the Gamma variant, seven point mutations in the Omicron BA.1 variant, and nine point mutations in the Omicron BA.2 variant. Furthermore, S-specific CD8+ T-cell responses were found more in convalescent individuals than in the vaccinated population.
Overall, the study findings highlighted the importance of mRNA vaccine-induced SARS-CoV-2 S-specific CD8+ S-specific T-cell responses against infections and disease progression of SARS-CoV-2 variants of concern.