SARS-CoV-2 escapes antiviral immunity by expressing viral proteins that disrupt signaling, detection, and interferon (IFN)-stimulated genes (ISGs) and IFN generation. A poor induction of type I IFN is linked to a hyperinflammatory response in severe CoV disease 2019 (COVID-19) patients. Insights into the mechanisms that inhibit SARS-CoV-2 multiplication and host inflammatory reactions are crucial for interpreting variance in COVID-19 severity and might lead to novel preventative and treatment options.
Previously, the authors of the present study discovered CNBP as a critical signaling molecule stimulated downstream of ribonucleic acid (RNA)-sensing pattern recognition receptors (PRRs). CNBP governed the transcription of type I IFNs to RNA and double-stranded RNA (dsRNA) viruses. CNBP normally resides in the cytosol. To switch on IFN gene transcription, CNBP endures nuclear translocation, phosphorylation, and IFN enhancer deoxyribonucleic acid (DNA) binding following RNA sensing mechanisms.
About the study
In the present study, the scientists uncovered the function of CNBP in limiting SARS-CoV-2 infection. Transfecting CNBP into angiotensin-converting enzyme 2 (ACE2) expressing adenocarcinomic human alveolar basal epithelial (A549) cells or vector control cells allowed the researchers to investigate the impact of CNBP in regulating COVID-19. They tracked the buildup of dsRNA employing J2 antibody immunofluorescence staining as a readout of viral infection.
SARS-CoV-2 concentrations and viral non-structural protein 14 (NSP14) and nucleocapsid protein (NP) levels in CNBP-expressing and vector control cells were analyzed using plaque assay. The team assessed the concentrations of SARS-CoV-2 proteins in CNBP-deficient A549-ACE2 cells following viral infection by measuring anti-NP antibodies.
The researchers tested whether the mutant of CNBP could still limit SARS-CoV-2 multiplication in transfected A549-ACE2 cells. They did this by transfecting the CNBP mutant (CNBP-M) and wild-type (WT) variants and tracking SARS-CoV-2 infection. In IFN a/b receptor (IFNAR) knock-out (KO) A549 ACE2 cells, A549-ACE2 cells without the IFNl receptor (IFNLR), and cells exposed to an anti-IFNAR antibody, the effect of CNBP overexpression in preventing SARS-CoV-2 replication was determined.
The team analyzed the mechanism via CNBP attach SARS-CoV-2 RNA by conducting RNA immunoprecipitation (RIP). Additionally, they performed a quantitative polymerase chain reaction (qPCR) to measure viral NSP-14 and NP RNA levels. Further, the regions of SARS-CoV-2 genomic RNA bound by CNBP were mapped. The researchers used WT littermate controls and CNBP deficient mice to analyze whether CNBP was significant in limiting SARS-CoV-2 in vivo. The effect of CNBP in controlling human CoV OC43 (HCoV-OC43) infection, another β-CoV, was also evaluated.
Results and discussions
The in vitro data of this study showed that SARS-CoV-2 generates poor and delayed ISGs and type I IFNs in infected cells relative to other viruses. REL-associated protein (RELA), CNBP, and IFN-regulatory factor 3 (IRF-3) demonstrated modest phosphorylation and nuclear translocation. These findings suggest that CNBP was under-stimulated in SARS-CoV-2-infected cells, possibly due to RNA sensors' inability to identify the virus and trigger downstream signaling.
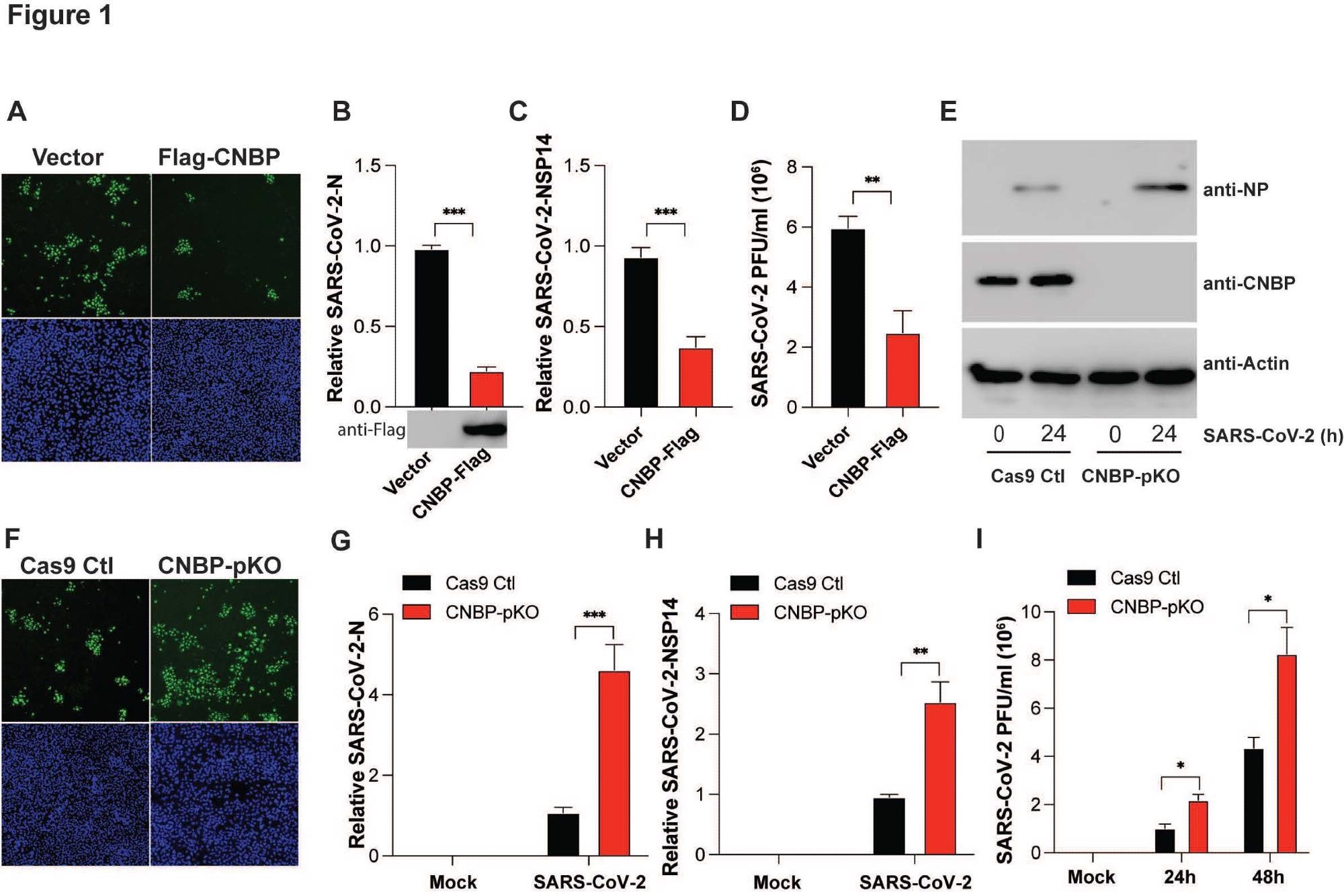 CNBP inhibits SARS-CoV-2 replication in vitro (A) hACE2-A549 cells were transfected with a Flag-CNBP expression plasmid or control, infected with SARS-CoV-2 for 24 hrs, and dsRNA was visualized by immunofluorescence with anti-J2 antibody (green). (B-D) Normalized SARS-CoV-2 RNA levels of NP (B) and NSP14 (C) as well as the SARS-CoV-2 titers (D) in hACE2-A549 cells transfected with Flag-CNBP plasmid and infected with SARS-CoV-2. (E-I) CNBP pKO and Cas9 Ctl A549 cells were infected with SARS CoV-2 at an MOI of 0.01. At 24 h post-infection, western blotting with viral NP protein expression (E), immunofluorescence staining with anti-J2 antibody (F), qPCR analysis of vRNA levels of NP (G) and NSP14 (H) as well as the viral titers assessed by plaque assay (I) in the supernatants were determined.
CNBP inhibits SARS-CoV-2 replication in vitro (A) hACE2-A549 cells were transfected with a Flag-CNBP expression plasmid or control, infected with SARS-CoV-2 for 24 hrs, and dsRNA was visualized by immunofluorescence with anti-J2 antibody (green). (B-D) Normalized SARS-CoV-2 RNA levels of NP (B) and NSP14 (C) as well as the SARS-CoV-2 titers (D) in hACE2-A549 cells transfected with Flag-CNBP plasmid and infected with SARS-CoV-2. (E-I) CNBP pKO and Cas9 Ctl A549 cells were infected with SARS CoV-2 at an MOI of 0.01. At 24 h post-infection, western blotting with viral NP protein expression (E), immunofluorescence staining with anti-J2 antibody (F), qPCR analysis of vRNA levels of NP (G) and NSP14 (H) as well as the viral titers assessed by plaque assay (I) in the supernatants were determined.
Due to inadequate RNA sensing, only a few CNBPs translocate to the nucleus to activate type I IFNs in SARS-CoV-2-infected cells. The majority of CNBP remains in the cytosol, where it can still stop SARS-CoV-2 replication. Furthermore, the current findings imply that CNBP works to limit viral replication in a cell-autonomic manner. The connection of the viral NP and genomic RNA, which results in higher-order RNA-protein compounds, was a significant process in SARS-CoV-2 multiplication, enabling the accumulation of RNA and proteins in virion assembly. CNBP performs this essential process via interfering phase separation between viral NP and RNA.
Mechanistically, CNBP attaches SARS-CoV-2 genomic RNA and prevents the NP from generating condensates. The 3′ untranslated region (3' UTR) and 5′ UTR were critical for the liquid-liquid phase separation (LLPS) seen with NP-RNAs, and CNBP binds both areas. As a result, the current findings showed that CNBP disturbs the NP's LLPS, highlighting the SARS-CoV-2 NP LLPS as a potential therapeutic option during COVID-19.
The scientists discovered that CNBP-deficient animals were more susceptible to SARS-CoV-2 infection, consistent with its role in modulating type I IFNs and effect on RNA-NP condensates. The influence of CNBP deficiency was more significant than that of IFNAR-deficient mice, highlighting CNBP's dual role. Additionally, the study revealed that CNBP positively controls type I IFN expression in RNA virus infection. Furthermore, the authors found that CNBP inhibited HCoV-OC43 replication through IFN-independent and dependent pathways similar to SARS-CoV-2.
Conclusions
The study findings illustrated that CNBP regulates IFNβ gene transcription in SARS-CoV-2-infected cells. Moreover, the authors found that CNBP links directly to SARS-CoV-2 RNA. CNBP contests with the NP and inhibits viral NP and RNA from undergoing LLPS assembling condensates vital for SARS-CoV-2 multiplication. Accordingly, animals and cells without CNBP exhibit higher SARS-CoV-2 loads, and CNBP-deficient mice die rapidly to infection.
Collectively, these inferences pinpoint CNBP as a crucial antiviral element for SARS-CoV-2, serving both as a cell-intrinsic limitation factor and a controller of antiviral IFN gene expression that interrupts LLPS to restrict viral spread and replication.

 *Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
Katherine Fitzgerald, Yongzhi Chen, Xuqiu Lei et al. CNBP restricts SARS-CoV2 by regulating IFN and disrupting RNA-protein condensates, 02 May 2022, PREPRINT (Version 1) available at Research Square [https://doi.org/10.21203/rs.3.rs-1576788/v1], https://www.researchsquare.com/article/rs-1576788/v1