Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections have been widely reported in individuals with a history of COVID-19 vaccination or infection with a preceding variant. This indicates that immunological heterogeneity exists against SARS-CoV-2 infection.
 Study: SARS-CoV-2 evolution and patient immunological history shape the breadth and potency of antibody-mediated immunity. Image Credit: NIAID
Study: SARS-CoV-2 evolution and patient immunological history shape the breadth and potency of antibody-mediated immunity. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
This study aimed to evaluate the potency and extent of antibodies produced following different immunological reactions against different variants of SARS-CoV-2.
The team obtained serum samples and compiled immunological histories assessed based on SARS-CoV-2 spike 1 (S1) proteins and spike-receptor binding domain (S-RBD) via enzyme-linked immunosorbent assay (ELISA). Metadata consisting of the date of serum sample collection, SARS-CoV-2 polymerase chain reaction (PCR) status, and vaccination status were also collected. The team further classified the serum samples into four groups, namely, naive (N), vaccinated (V), infected (I), and infected and vaccinated (IV). The serum samples from the I and IV groups were further sub-classified based on the infecting SARS-CoV-2 variant as infected-Wuhan (IW), infected-alpha (Iα, 39 samples), 107 infected-delta (Iδ, 15 samples), infected-Wuhan-vaccinated (IWV, 60 samples), infected-alpha vaccinated (IαV, 69 samples), and infected-delta-vaccinated (IδV).
The serum samples were then processed by testing them against the S and nucleocapsid (N) protein using an electrochemiluminescence assay to quantify the antibody concentrations. Virus neutralization assays (VNAs) were also performed on the serum samples using human immunodeficiency virus (HIV) pseudotypes that carried the S protein for either the SARS-CoV-2 Wuhan, Alpha, Delta, or Omicron strains.
Utilizing viral pseudotypes, they also evaluated whether each serological sample neutralized the four SARS-CoV-2 strains. Furthermore, the neutralization ability of the antibody-mediated response was quantified among the N, V, I, and IV groups by titrating the neutralizing antibodies against each SARS-CoV-2 variant.
Results
The study results showed that the serum samples collected from the naive patients had the lowest levels of anti-SARS-CoV-2 S antibodies since these participants were not exposed to the SARS-CoV-2 S antigen. S antibody levels were higher in infected individuals than in vaccinated individuals, while the levels were significantly lower in participants who had both been infected and vaccinated. Furthermore, participants who were infected had greater amounts of anti-SARS-CoV-2 N antibodies in comparison to those who had been infected and vaccinated.
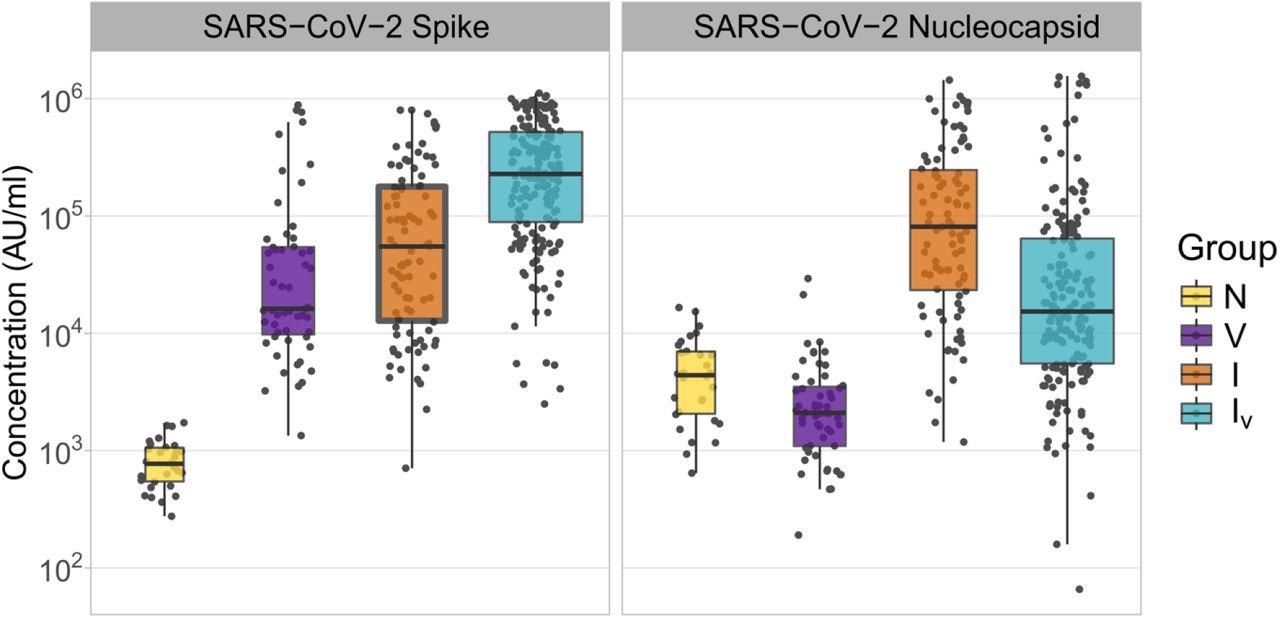
Concentrations of SARS-CoV-2 Spike and Nucleocapsid antibodies in samples derived from patients with different histories of SARS-CoV-2 exposure. The name of the antigens is shown at the top of each panel. Patient groups are defined as N: naïve (yellow); V: vaccinated (purple); I: infected (orange); IV: infected and vaccinated (cyan). Antibody concentrations are shown in MSD arbitrary units/ml. Boxplots displayed the interquartile range and median values.
Notably, the anti-N levels in the vaccinated persons were significantly lower than those in the naive individuals. Overall, these findings indicated that exposure to SARS-CoV-2 antigens by both vaccination and infection resulted in higher amounts of anti-SARS-CoV-2 antibodies as compared to those after only vaccination.
The team found that the neutralization activity of each sample was dependent on the infecting SARS-CoV-2 variant and the immunological history of the respective patient. A consistent reduction in neutralization and antigenic drift could be identified by considering the chronology in which the variants appeared.
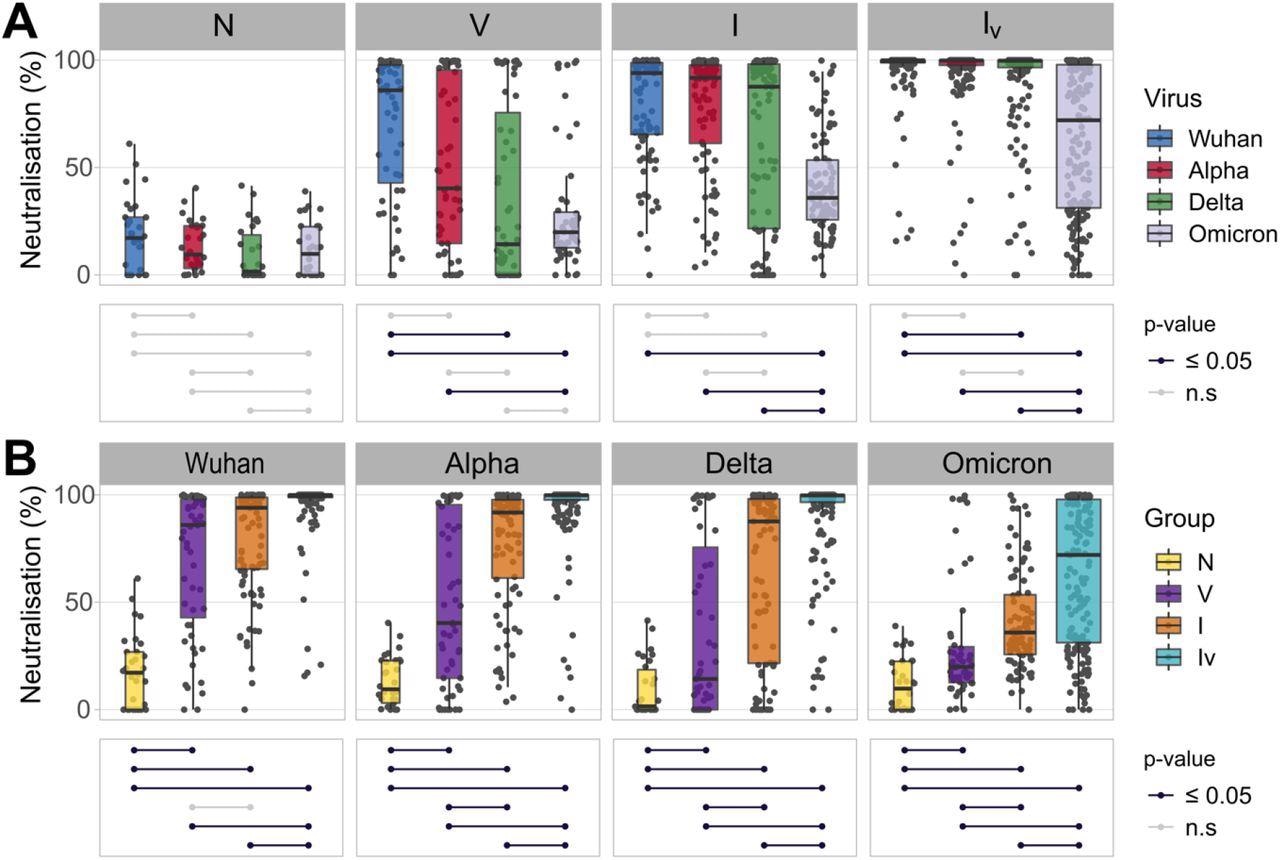
Neutralizing responses elicited against pseudotyped viruses carrying the S protein of different SARS-CoV-2 variants according to patient exposure history to SARS-CoV-2. (A) Sera from patients were grouped based on immunological histories (N: naive; V: vaccinated; I: infected; IV: infected and vaccinated). Neutralizing activity was measured using Wuhan (blue), Alpha (red), Delta (green) and Omicron (grey) spike glycoprotein bearing HIV(SARS-CoV-2) pseudotypes and plotted per patient group (A) and per SARS-CoV-2 S variant (B). Neutralization was measured at a fixed dilution (1:50). Each point represents the mean of two replicates. Boxplots displayed the interquartile range and median values. Significance levels between patient groups or pseudotyped viruses were tested using the pairwise Wilcoxon test, and are shown in bottom panels as connected dot plots.
Moreover, patients belonging to the IV group showed higher neutralizing antibody titers than those found in all the other groups corresponding to each variant tested. On the other hand, infected patients displayed variable antibody titers against each SARS-CoV-2 variant as compared to the vaccinated participants. This was noted in the insignificant differences observed in the infected and vaccinated groups corresponding to the SARS-CoV-2 Wuhan, Alpha, Delta, and Omicron variants. However, vaccinated patients had substantially higher antibody titers against Omicron while the neutralization potency was very low. Overall, this indicated that the evolution of SARS-CoV-2 antigens is directional, that is, it has evolved to evade antibody-mediated immunity, and that the type and number of viral exposures affect the potency of antibody-mediated immunity.
The team also noted that patients who were both vaccinated and had a history of Delta infection showed the highest neutralization potency against the rest of the variants. Based on neutralization bias measurements, individuals infected with specific variants had distinct antibody titers against other variants. This indicated that patients belonging to the infected group had antibody titers lower than those who were both vaccinated and infected. In contrast, the neutralizing antibody titers of patients infected with the Delta variant against the homologous antigen were similar to those in vaccinated and infected individuals.
Furthermore, the comparison of neutralizing antibody titers across all patient groups showed that individuals who were vaccinated before being infected with the Delta variant or those that were infected with the Wuhan variant before receiving their vaccination had the highest antibody titers against all the SARS-CoV-2 variants. This indicated that the patients who were infected before being vaccinated had higher neutralization potency and antibody titers against each variant.
Conclusion
The study findings highlighted the complexity of the impact of SARS-CoV-2 infection on human immunity. Researchers believe the current study will improve epidemiological models in order to forecast future SARS-CoV-2 transmission trends. In addition, further studies are needed to understand the effects of prior SARS-CoV-2 exposure on the manifestation and outcome of COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
SARS-CoV-2 evolution and patient immunological history shape the breadth and potency of antibody-mediated immunity. Maria Manali, Laura A Bissett, Julien Amat, Nicola Logan, Sam Scott, Ellen Hughes, William Harvey, Richard Orton, Emma Thomson, Rory Gunson, Mafalda Viana, Brian Willett, Pablo Ramiro Murcia, bioRxiv 2022.05.06.490867, DOI: https://doi.org/10.1101/2022.05.06.490867, https://www.biorxiv.org/content/10.1101/2022.05.06.490867v1
- Peer reviewed and published scientific report.
Manali, Maria, Laura A Bissett, Julien A R Amat, Nicola Logan, Sam Scott, Ellen C Hughes, William T Harvey, et al. 2022. “SARS-CoV-2 Evolution and Patient Immunological History Shape the Breadth and Potency of Antibody-Mediated Immunity.” The Journal of Infectious Diseases 227 (1): 40–49. https://doi.org/10.1093/infdis/jiac332. https://academic.oup.com/jid/article/227/1/40/6653591.