The ongoing coronavirus disease 2019 (COVID-19) pandemic caused by SARS-CoV-2 has led to over 524 million cases and six million mortalities globally. Intensive worldwide efforts are continuing to develop, analyze, and adopt vaccinations or other medical alternatives against COVID-19, such as monoclonal antibody (mAb) treatment. However, SARS-CoV-2 variants that evade neutralization imparted by COVID-19 therapeutic antibodies and vaccination/infection acquired immunity have emerged because of widespread viral transmission and crucial mutations.
Notably, most presently approved COVID-19 therapeutic mAbs have exhibited decreased neutralizing effectiveness against the SARS-CoV-2 Omicron variant and its sublineages, like BA.2 and BA.1, due to approximately 30 mutations in its spike (S) protein. This emphasizes the persistent need to discover mAbs that are effective against new SARS-CoV-2 mutants.
About the study
The authors of the present research have previously assessed humoral immunity in 42 COVID-19 convalescent individuals who experienced mild symptoms following the transmission of the SARS-CoV-2 ancestral Wuhan strain (WA.1) in the year 2020.
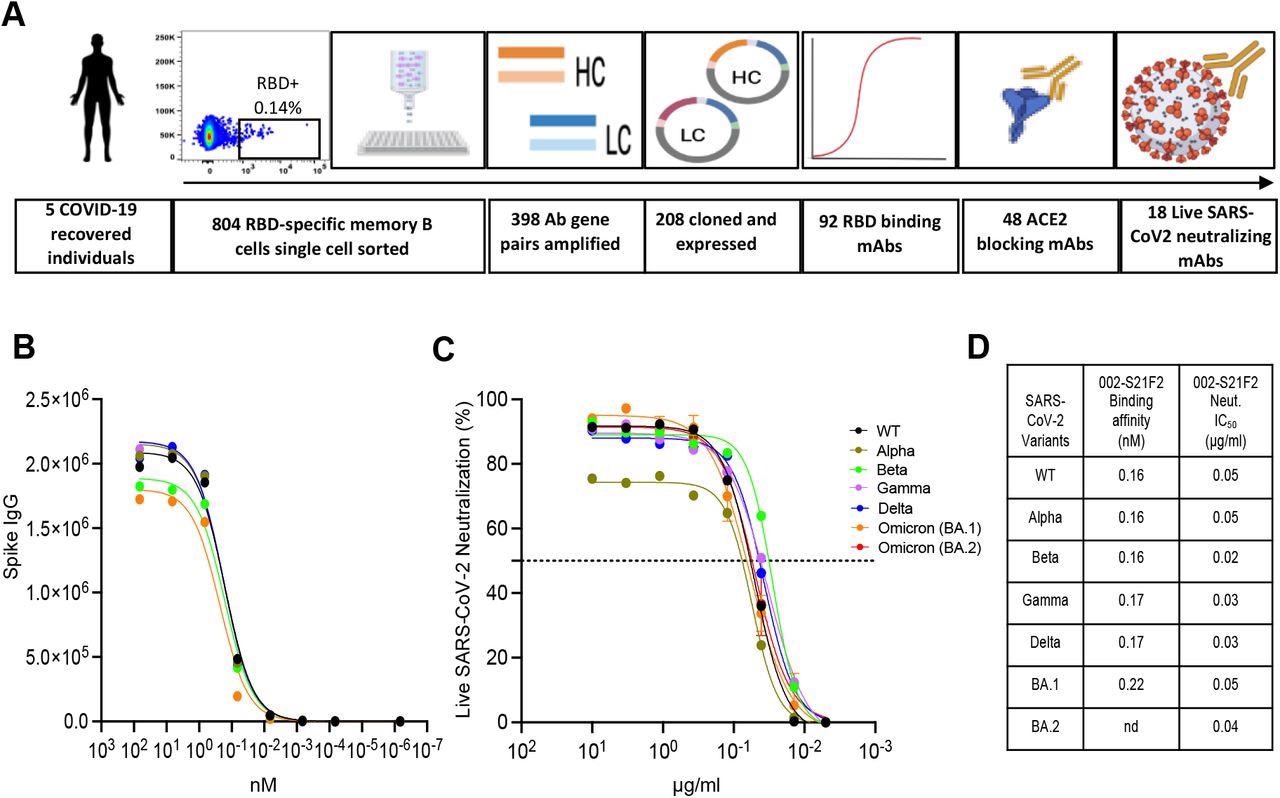
Identification of a broad and potent SARS-CoV-2 RBD specific human monoclonal antibody 002-S21F2. (A) The overall strategy for the isolation of RBD-specific mAbs described in this study. (B) 002-S21F2 was tested for binding to the spike proteins of SARS-CoV-2 WA.1, Alpha, Beta, Gamma, Delta and Omicron variants of concern (VOC). (C) Authentic live virus neutralization curves of 002-S21F2 for WA.1, Alpha, Beta, Gamma, Delta and Omicron (BA.1 and BA.2) SARS-CoV-2 VOCs. Neutralization was determined on Vero-TMPRSS2 cells using a focus reduction assay. (D) 002-S21F2 mediated neutralization 50% inhibitory concentration IC50 values were obtained from live SARS-CoV-2 VOC neutralization assays. Affinity constant (KD) values calculated from the binding curves for two mAbs as measured by the MSD binding assays are plotted.
In the present study, the researchers selected five subjects from the prior research with measurable frequencies of memory B cells specific to the SARS-CoV-2 receptor-binding domain (RBD), high RBD binding concentrations, and substantial neutralization levels to live SARS-CoV-2 WA.1 strain for the production of SARS-CoV-2 RBD-targeting mAbs.
The team screened 804 class-switched B cells engaging with SARS-CoV-2 RBD fluorescent probe and amplified 398 coupled light- and heavy-chain antibody gene sequences. Further, they conducted enzyme-linked immunosorbent assay (ELISA) profiling in the selected mAbs.
The scientists assessed the binding capacities of the 002-S21F2 antibody towards several SARS-CoV-2 variants of concern (VOCs) S protein via an electrochemiluminescence multiplex experiment. They used cryogenic electron microscopy (cryo-EM) structures of 002-S21F2 full-length immunoglobulin G (IgG) complexed with Omicron and WA.1 S proteins to delineate the molecular characteristics and mechanisms of the broad SARS-CoV-2 neutralization spectrum of 002-S21F2.
Results
The study results indicated that the authors successfully cloned and produced 208 SARS-CoV-2 RBD-targeting mAbs from the 398 amplified matched light- and heavy-chain antibody sequences. Additionally, the ELISA screening resulted in 92 SARS-CoV-2 RBD-binding mAbs. The majority of these mAbs displayed a poor rate of somatic hypermutations (SHM) in their light and heavy chains, indicating they were recently generated from a naïve B cell population. Among these antibodies, 48 mAbs successfully prevented host angiotensin-converting enzyme 2 (ACE2) and viral RBD contact, and 18 effectively neutralized live virus with half-maximal inhibitory concentrations (IC50) varying from 0.05 to 17 µg/ml.

Antibody 002-S21F2 exhibits distinct genetic and epitope contact features in comparison to SARS-CoV-2 therapeutic antibodies. (A) Comparison of 002-S21F2 mAb genetic feature with therapeutic mAbs in clinics. Omicron neutralizing mAbs are highlighted in bold and red color (B) Comparison of 002-S21F2 (green) epitope site with S309 (Sotrovimab) (red outline), Ly-CoV1404 (Bebtelovimab) (yellow outline) epitopes on SARS-CoV-2 RBD.
The authors noted that among all the mAbs that neutralized live SARS-CoV-2 WA.1 sequence, antibody 002-S21F2 was the most effective. They found that 002-S21F2 adhered to all analyzed SARS-CoV-2 variant S proteins with equal affinities. Besides, 002-S21F2 bonded to prefusion-stabilized WA.1 S exhibiting picomolar affinity according to biolayer interferometry. In addition, the SARS-CoV-2 RBD addressing human antibody, 002-S21F2, efficiently neutralized live viral isolates of SARS-CoV-2 VOCs, including Beta, Alpha, Delta, Gamma, and Omicron BA.2 and BA.1 with IC50 spanning from 0.02 to 0.05 ug/ml.
This near germline antibody harbored genetic characteristics that set it apart from other SARS-CoV-2 mAbs. 002-S21F2 identified an epitope on the external surface of RBD, i.e., class-3 surface, according to cryo-EM structural investigations. Furthermore, this epitope was outside of the ACE2 engaging motif, and its distinct molecular properties allowed it to overcome mutations observed in the Omicron variants.
The antigenic residues addressed by the broadly neutralizing antibody (bnAb), 002-S21F2, were significantly conserved between previous and present SARS-CoV-2 VOCs. The structural data revealed that 002-S21F2 maintained effective neutralization against Omicron BA.2 and BA.1 variants possessing epitope mutations at N440K and G339D. Since the recently listed Omicron BA.2.12.1, BA.2.13, BA.5, BA.4, and BA.3 variants were not known to possess any additional changes within the 002-S21F2 epitope area, the scientists assumed that this mAb's neutralizing ability would endure against them. Additionally, sequence alignment of Sarbecovirus RBDs demonstrated that 10 of 19 conserved residues of SARS-CoV were addressed by 002-S21F2, indicating the possibility of cross-reactivity with other Sarbecoviruses.
Structural comparison of other class-3 mAbs, such as the two current therapeutic mAbs targeting Omicron, Bebtelovimab (Ly-CoV1404) and Sotrovimab (S309), and the 002-S21F2 epitope demonstrated some similarities among the C135 and 002-S21F binding sites. Nonetheless, C135 did not neutralize the Omicron VOC.
Furthermore, the structural analysis revealed that the small number of SHMs found in this near germline 002-S21F2 bnAb were not implicated in antigenic site recognition, suggesting that the footprints of such bnAbs might offer a template to drive rational COVID-19 vaccine development.
Overall, the authors mentioned that the identification and detailed structural characterization of 002-S21F2 in this study provided important information regarding their broad and effective neutralization of SARS-CoV-2 VOCs.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Structural insights for neutralization of BA.1 and BA.2 Omicron variants by a broadly neutralizing SARS-CoV-2 antibody. Sanjeev Kumar, Anamika Patel, Lilin Lai, Chennareddy Chakravarthy, Rajesh Valanparambil, Meredith E. Davis-Gardner, Venkata Viswanadh Edara, Susanne Linderman, Elluri Seetharami Reddy, Kamalvishnu Gottimukkala, Kaustuv Nayak, Prashant Bajpai, Vanshika Singh, Filipp Frank, Narayanaiah Cheedarla, Hans Verkerke, Andrew S. Neish, John D. Roback, Grace Mantus, Pawan Kumar Goel, Manju Rahi, Carl W. Davis, Jens Wrammert, Mehul S. Suthar, Rafi Ahmed, Eric Ortlund, Amit Sharma, Kaja Murali Krishna, Anmol Chandele. bioRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.05.13.491770, https://www.biorxiv.org/content/10.1101/2022.05.13.491770v1
- Peer reviewed and published scientific report.
Kumar, Sanjeev, Anamika Patel, Lilin Lai, Chennareddy Chakravarthy, Rajesh Valanparambil, Elluri Seetharami Reddy, Kamalvishnu Gottimukkala, et al. 2022. “Structural Insights for Neutralization of Omicron Variants BA.1, BA.2, BA.4, and BA.5 by a Broadly Neutralizing SARS-CoV-2 Antibody.” Science Advances 8 (40). https://doi.org/10.1126/sciadv.add2032. https://www.science.org/doi/10.1126/sciadv.add2032.