There are several similarities between COVID-19-induced ARDS and non-COVID-19-induced ARDS in terms of clinical presentations and pathologies. However, COVID-19 ARDS is characterized by a more potent inflammatory response than traditional ARDS.
 Study: Multi-omic comparative analysis of COVID-19 and bacterial sepsis-induced ARDS. Image Credit: Chinnapong / Shutterstock
Study: Multi-omic comparative analysis of COVID-19 and bacterial sepsis-induced ARDS. Image Credit: Chinnapong / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
In the present study, researchers conducted a multi-omic analysis to identify and compare signatures found in COVID-19 ARDS and non-COVID-19 ARDS based on the subnetworks.
The team assessed the molecular variations between bacterial sepsis-induced and COVID-19-induced ARDS by analyzing three molecular layers, including metabolic, lipidomic, and proteomic. The biological processes responsible for the variations between COVID-19- and bacterial sepsis-induced ARDS were identified by annotating the differentiated molecules. This molecular annotation was performed using Metabolon’s sub-pathways wherein the lipids were annotated using lipid classes while the proteins were annotated using the Kyoto encyclopedia of genes and genomes (KEGG) signaling pathways.
Furthermore, the team utilized previous studies that compared COVID-19 ARDS and non-COVID-19 ARDS with either less severe COVID-19 infections or healthy controls. Moreover, the team selected processes associated with ARDS to investigate on a molecular level.
The variations in omics correlations with clinical manifestations were also assessed across the two ARDS cohorts. These evaluations included the assessment of acute kidney injury (AKI), PaO2/FiO2 ratio, thrombocytosis, and mortality. A systemic view of the multi-omic molecules in the two ARDS groups was developed via a Gaussian graphical model that identified the interactions incident between the molecules.
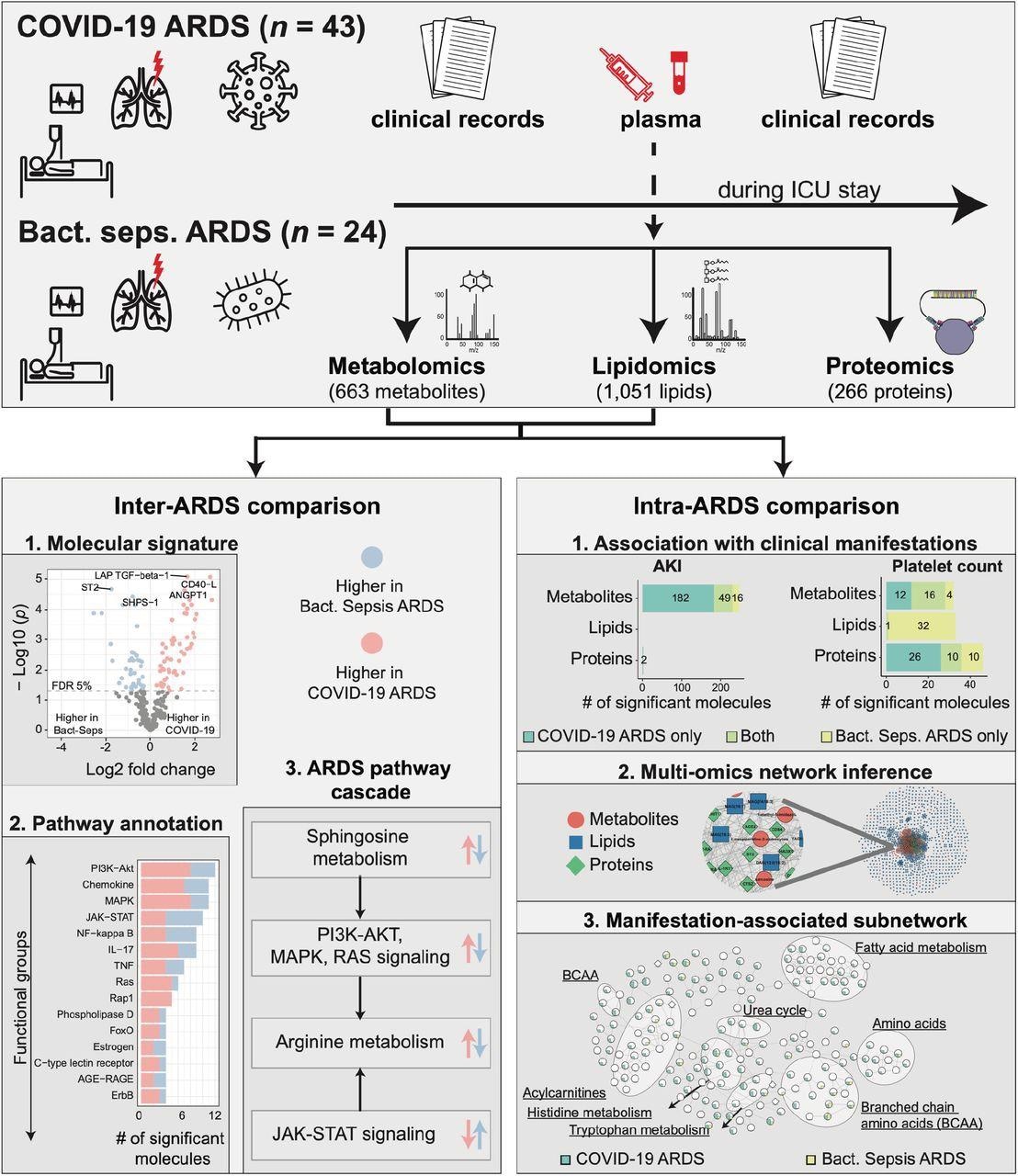
Study overview. This study was based on 67 ARDS patients, 43 with COVID-19 and 24 with bacterial sepsis group. Profiling of plasma samples resulted in 1,906 measured molecules, including 663 metabolites, 1,051 lipids, and 266 proteins. For inter-ARDS comparison, we identified molecules and pathways differently regulated between the two ARDS groups. In addition, focusing on several selected pathways with therapeutic relevance, we constructed a cascade of biological processes starting from sphingosine metabolism. For intra-ARDS comparison, we identified molecules associated with clinical manifestations, including acute kidney injury (AKI), thrombocytosis (platelet count), PaO2/FiO2 ratio, and mortality, within each ARDS group. Further, we constructed a data-driven multi-omic network based on the Gaussian graphical model (GGM). This network was used to generate subnetworks associated with clinical manifestations.
Results
The team analyzed a total of 67 patients who were admitted to the intensive care unit (ICU), including 24 patients diagnosed with bacterial sepsis and 43 with COVID-19. Among these patients, 25.4% were women with a median age of 60 years. Approximately 11 of the COVID-19 ARDS and nine bacterial sepsis-induced ARDS patients succumbed. Moreover, almost 46.3% of the patients reported acute kidney injury (AKI), including 16 in the COVID-19 ARDS and 15 in the bacterial sepsis-induced ARDS cohorts.
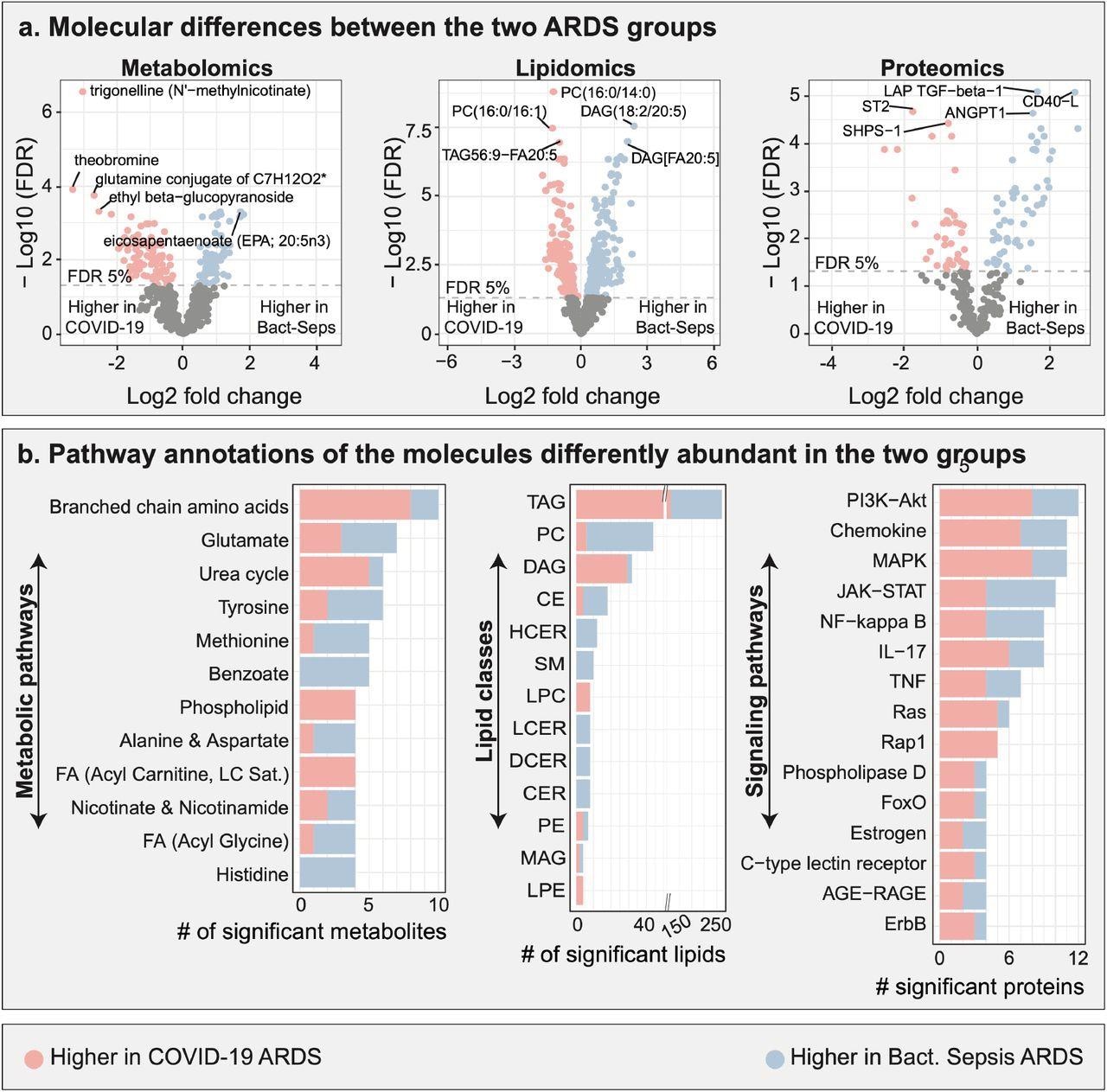
Multi-omic comparison between COVID-19 ARDS and bacterial sepsis-induced ARDS. a. Metabolomic, lipidomic, and proteomic analyses between the two ARDS groups. 706 molecules were differently abundant in the two ARDS groups. b. Functional annotations of significant molecules. Pathways and classes with at least 4 significant molecules were included in these plots. FDR – false discovery rate. Lipid class abbreviations: TAG – Triacylglycerol, PC – Phosphatidylcholine, DAG – Diacylglycerol, CE – Cholesteryl ester, HCER – Hexosylceramides, Total – total lipids, SM – Sphingomyelin, LPC – Lysophosphatidylcholine, LCER - Lactosylceramides, DCER – Dihydroceramides, CER – Ceramides, PE – Phosphatidylethanolamine, MAG – Monoacylglycerol, LPE - Lysophosphatidylethanolamine, PI - Phosphatidylinositol.
The plasma molecular profile analysis showed that 175 of the 663 metabolites, 437 of the 1051 lipids, and 94 of the 266 proteins assessed were differentially abundant among the plasma groups. The team also observed that 10 metabolites belonging to the branched-chain amino acids (BCAAs) pathway were found in abundance between the two ARDS groups. Among these, eight displayed higher, and two displayed lower levels of COVID-19-induced ARDS in comparison to those in bacterial sepsis-induced ARDS.
Seven metabolites from the glutamate metabolism pathway were differentially abundant between the two ARDS groups, including four metabolites that had higher and three that had lower levels in the COVID-19-induced ARDS samples than in the bacterial sepsis-induced ARDS samples. The team also found significant changes in the lipidomic profiles between the ARDS groups with the most differences observed in the triacylglycerols (TAGs) and diacylglycerols (DAGs) lipid classes. This underlined the role of lipid metabolism in disease prognosis. The researchers found that the amounts of TAGs and DAGs were higher in COVID-19 ARDS than in bacterial sepsis-induced ARDS.
A total of 12 proteins from the phosphoinositide-3 kinase-AKT signaling pathway were abundant between the two ARDS, including eight having higher and four having lower levels in COVID-19 ARDS as compared to bacterial sepsis-induced ARDS. On the other hand, 11 proteins belonging to the mitogen-activated protein kinase (MAPK) signaling pathway were abundant between the two ARDS groups, including eight that had higher and three that had lower levels in COVID-19 ARDS as compared to bacterial sepsis-induced ARDS.
In omics analysis, significant associations were found concerning the PaO2/FiO2 ratio in the COVID-19 ARDS cohort, while none of the molecules were correlated with mortality. Moreover, 249 and 111 molecules were found to be associated with AKI and platelet count, respectively. This included 76 molecules that overlapped between the COVID-19 ARDS and bacterial sepsis-induced ARDS.
With respect to the AKI signature in COVID-19 ARDS patients, a total of 233 molecules were correlated with AKI, including 135 metabolites and two proteins that were positively associated with AKI, 96 metabolites that were negatively associated, while none of the lipids had any association. In bacterial sepsis-induced ARDS, 46 metabolites were positively and 19 metabolites were negatively associated with AKI, while no lipids or proteins displayed any correlation to AKI.
Overall, the study findings showed that multi-omic analysis could efficiently identify novel therapeutic approaches and establish different pathophysiological characteristics within the ARDS.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Multi-omic comparative analysis of COVID-19 and bacterial sepsis-induced ARDS, Richa Batra, William Wahlen, Sergio Alvarez-Mulett, Katherine Hoffman, Will Simmons, John Harrington, Kelsey Chetnik, Mustafa Buyukozkan, Elisa Benedetti, Mary E. Choi, Karsten Suhre, Frank Schmidt, Edward Schenck, Augustine M.K. Choi, Soo Jung Cho, Jan Krumsiek, medRxiv 2022.05.16.22274587, DOI: https://doi.org/10.1101/2022.05.16.22274587, https://www.medrxiv.org/content/10.1101/2022.05.16.22274587v1
- Peer reviewed and published scientific report.
Batra, Richa, William Whalen, Sergio Alvarez-Mulett, Luis G. Gomez-Escobar, Katherine L. Hoffman, Will Simmons, John Harrington, et al. 2022. “Multi-Omic Comparative Analysis of COVID-19 and Bacterial Sepsis-Induced ARDS.” Edited by Jacob S. Yount. PLOS Pathogens 18 (9): e1010819. https://doi.org/10.1371/journal.ppat.1010819. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1010819.