A recent article under review at the Nature Portfolio journal and currently available on the Research Square* preprint server analyzed the factors influencing the durability of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epitope-specific T cell reaction.
The ongoing coronavirus disease 2019 (COVID-19) pandemic caused by SARS-CoV-2 has precipitated substantial mortality and morbidity globally. It is well known that the generation of neutralizing antibodies and a T cell response towards the SARS-CoV-2 antigen drives the recovery from COVID-19.
Studies indicated that T cells harbor a vital function in lowering illness severity during COVID-19 and establishing long-standing immune memory. However, what factors determine the durability of the SARS-CoV-2-selective T cell reaction is one of the main concerns. An in-depth examination of T cell reaction characteristics at individual clones and epitopes level could reveal elements that influence the persistence and development of long-lasting T cell memory.
About the study
In the present work, the scientists assessed humoral and T cell reactions to SARS-CoV-2 in matched blood samples from 50 COVID-19 convalescent patients (CPs) promptly after infection and at an average of eight months following COVID-19. They also studied the prevalence of memory T cells, their clonal architecture, and the T-cell receptor (TCR) repertoire of the T cell immune reaction to an array of eight CD8+ epitopes in a subset of 26 CPs. The present study assessed CD8+ T cell reactivity to specific epitopes at the clonotype degree.
The 50 SARS-CoV-2 CPs had asymptomatic, mild, or moderate to severe COVID-19 based on the United States (US) National Institutes of Health's classification. The authors assessed the presence of immunoglobulin Gs (IgGs) targeting the SARS-CoV-2 receptor-binding domain (RBD) in all participants. Further, they studied the T cell reactions to peptide collections obtained from the SARS-CoV-2 spike (S), membrane (M), and nucleoprotein (N) proteins.
The researchers chose 20 CD8+ epitopes displayed by frequent human leukocyte antigen I (HLA I) alleles and generated from various SARS-CoV-2 proteins that had previously been reported as immunogenic to analyze T cell response at individual epitope levels. Additionally, using high-throughput sequencing, they examined the repertoires of TCR-beta chains in major histocompatibility complex (MHC)-tetramer+ groups and entire peripheral blood mononuclear cell (PBMC) portions.
Results and discussions
The scientists discovered that T cell responses were produced more frequently and lasted longer relative to circulating antibodies in a larger cohort of recovered SARS-CoV-2 patients. Nonetheless, cellular immunity also deteriorated over time, as evidenced by a decline in the number of recognized epitopes and antigens, a lower prevalence of particular T cells in circulation, and a decreased number of clonotypes specific to the epitope. Yet, most epitopes were detected around 10 months after infection.
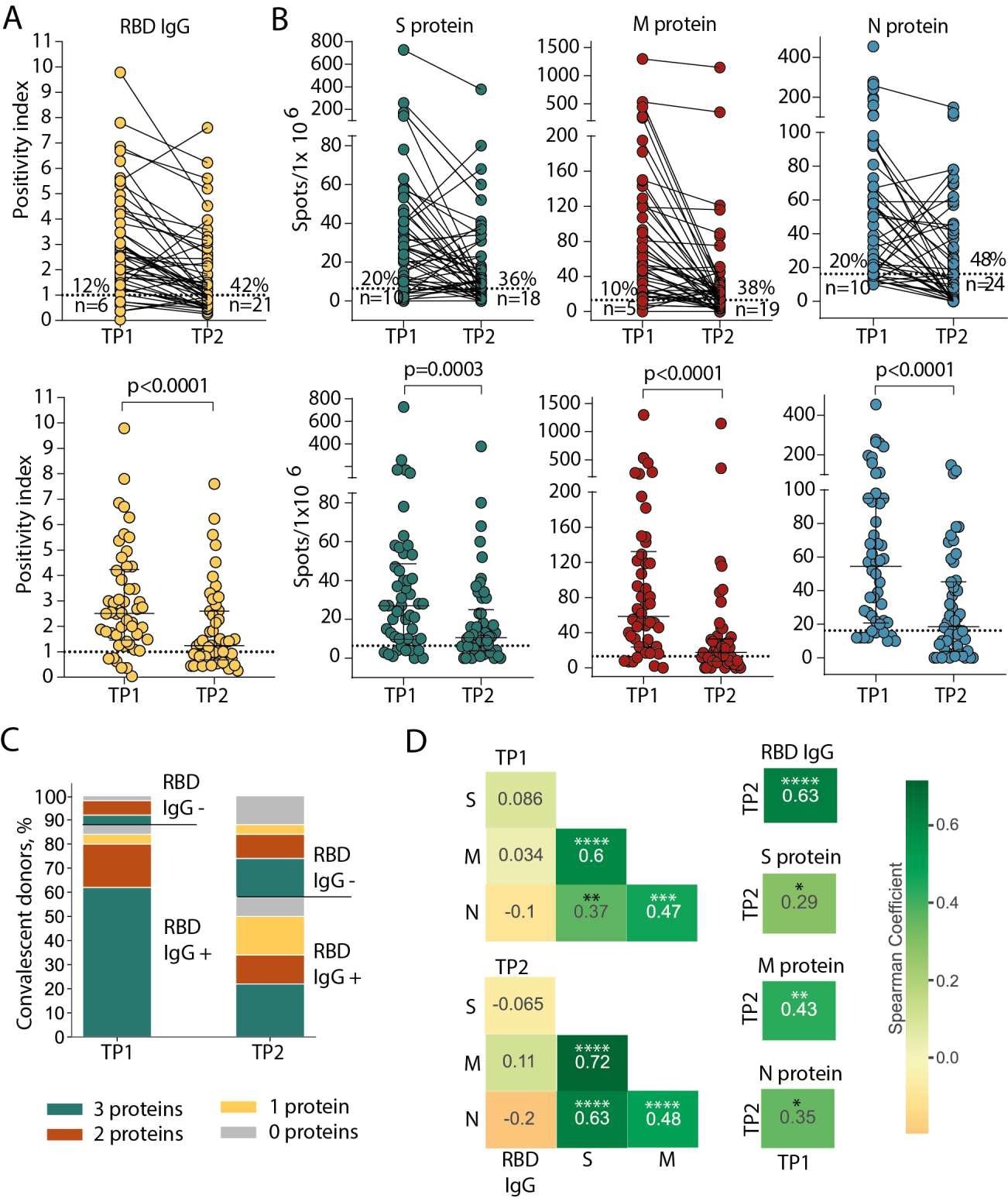
Persistence of humoral and cellular response in CP cohort. (A) Levels of anti-RBD IgG as measured by ELISA at the two time-points. Means of two independent measurements are plotted. Upper plot shows samples from the same donor, connected by lines. Lower plot shows median with interquartile range (paired Wilcoxon test). (B) Magnitude of T cell response to pools of peptides derived from S (green), M (red), and N proteins (blue) as measured by IFNγ ELISpot. Means of two independent measurements with negative control subtracted are plotted. Upper and lower plots are presented as in (A). Dotted lines in (A) and (B) mark cut-offs, the number and share of individuals without detectable response is indicated. N = 50 for both (A) and (B). (C). Distribution of immune responses in CPs. Colors indicate T cell response to 0–3 peptide pools; RBD IgG+ and RBD IgG- respectively indicate presence and absence of anti-RBD IgG. (D). Spearman correlation between humoral and cellular responses to different SARS-CoV-2 antigens. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
Among the 15 tested epitopes, the previously described epitopes (ATS, LLLLD, and NRF) were not immunogenic in the current investigation. The immunogenicity of the remaining epitopes varied. Indeed, 11 were immunodominant, eliciting a reaction in more than half of the CPs. In a healthy donor, just one epitope, i.e., LLY, elicited a response.
The amount of the shift in T cell response was not consistent throughout epitopes, with some epitopes sustaining significant immunogenicity for up to eight months. The investigators found that antigen-specific T cells generated persistent memory populations in blood and multiplied after being stimulated with their antigen. The present data proved that more ample clonotypes do not always persist longer. In addition, clonal disappearance and contraction were more associated with a highly proliferative phenotype.
The team discovered 756 distinct clonotypes for nine CD8+ T cell epitopes. Highly comparable public clonotypes identified some epitopes. Other epitope receptors were quite varied, implying alternate ways of recognition. TCRs with the highest mutual resemblance recognized LLY, accounting for eight of the 19 available CDR3-beta sequences, whereas the remaining epitopes showed modest degrees of homology to particular TCRs.
In peripheral blood, most epitope-specific clonotypes were found at quantities below the detection limit. The most significant variable impacting the prevalence of response to a specific epitope and its durability was the number of particular clonotypes discovered following infection. On the other hand, the average size of the clonotypes had no effect.
The number of identified epitopes for each patient and the proportion of epitope-specific clonotypes reduced over time. However, the number of specific T cells declined unevenly for the epitopes analyzed. Epitopes with a higher clonally varied TCR repertoire elicited stronger and longer-lasting responses. The excess of particular clonotypes in peripheral circulation, on the other hand, did not affect their longevity.
Conclusions
Overall, the study findings demonstrated that the clonal variety of the initial T cell response, instead of the prevalence of a specific clonotype in the peripheral circulation, was critical for establishing a long-lasting immune response to SARS-CoV-2.
In the light of the emergence of novel SARS-CoV-2 variants evading neutralizing antibodies, cellular immunity has become increasingly relevant in decreasing COVID-19 severity. To date, the most commonly used SARS-CoV-2 vaccines were centered on the S protein. On the contrary, only three of nine epitopes identified as immunodominant in this study were S-derived.
On the whole, the current work offers a rationale for incorporating immunodominant T cell epitopes identified by distinct T cell populations in the new generation of COVID-19 vaccines to assure the establishment of long-living T cell memory.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ksenia Zornikova, Alexandra Khmelevskaya, Savely Sheetikov et al. Clonal diversity determines persistence of SARS-CoV-2 epitope-specific T cell response., 17 June 2022, PREPRINT (Version 1) available at Research Square, DOI: https://doi.org/10.21203/rs.3.rs-1719448/v1, https://www.researchsquare.com/article/rs-1719448/v1
- Peer reviewed and published scientific report.
Zornikova, Ksenia V., Alexandra Khmelevskaya, Savely A. Sheetikov, Dmitry O. Kiryukhin, Olga V. Shcherbakova, Aleksei Titov, Ivan V. Zvyagin, and Grigory A. Efimov. 2022. “Clonal Diversity Predicts Persistence of SARS-CoV-2 Epitope-Specific T-Cell Response.” Communications Biology 5 (1): 1–11. https://doi.org/10.1038/s42003-022-04250-7. https://www.nature.com/articles/s42003-022-04250-7.