They used Immunome, Inc's discovery platform to investigate the memory B cell repertoires of three convalescent coronavirus disease 2019 (COVID-19) patients to derive IMM-BCP-01. The antibody cocktail performed well under in vitro conditions against multiple SARS-CoV-2 variants, including Omicron BA.1 and BA.2. When evaluated in vivo in golden Syrian hamsters, IMM-BCP-01 potently induced anti-SARS-CoV-2 effector response, including antibody-dependent cellular cytotoxicity (ADCC), complement pathway activation, and phagocytosis.
 Study: IMM-BCP-01, a patient-derived anti-SARS-CoV-2 antibody cocktail, is active across variants of concern including Omicron BA.1 and BA.2. Image Credit: Lightspring / Shutterstock
Study: IMM-BCP-01, a patient-derived anti-SARS-CoV-2 antibody cocktail, is active across variants of concern including Omicron BA.1 and BA.2. Image Credit: Lightspring / Shutterstock
Background
To date, the use of convalescent plasma against SARS-CoV-2 has fetched mixed results. Yet, antibody therapies with broad reactivity could serve as an alternative to COVID-19 vaccination. The emergence of Omicron sub-variants, with exceptional immune evasion properties, led the United States Food and Drug Administration (FDA) to revise the emergency use authorization (EUA) issued for some antibody cocktail treatments (e.g., Evusheld). The FDA also recommended increasing its dosage due to loss of potency to Omicron BA.1 and BA.1.1 sub-variants.
However, preliminary findings from therapies deploying a single antibody necessitated the use of cocktail treatment to prevent the generation of escape mutants. Therefore, there is an unmet urgent need to develop an antibody cocktail with broad reactivity against current and future SARS-CoV-2 variants.
About the study
In the present study, researchers derived a three antibody cocktail, the IMM-BCP-01, and characterized it based on each constituting antibody’s function, epitopes, and biochemical properties. They evaluated it using neutralization assays against current SARS-CoV-2 variants of concern (VOCs), effector functional assays in vitro, and in hamsters infected with SARS-CoV-2 variants in vivo.
Structural studies focused on the antibody interactions with SARS-CoV-2 spike (S) trimer protein. The team confirmed these findings by cryogenic electron microscopy (cryo-EM) analysis. Further, they used an alanine-scanning shotgun mutagenesis approach to validate the structural data; that used a validated library of the receptor-binding domain (RBD) proteins from the ancestral SARS-CoV-2 Wuhan-Hu 1 strain, each containing one amino acid mutation.
They also evaluated the pharmacokinetics (PK) profile of a three antibody cocktail, IMM-BCP-01. They injected naiive hamsters with IMM-BCP-01 and measured their total immunoglobulin G (IgG) serum levels using an enzyme-linked immunosorbent assay (ELISA). The IMM-BCP-01 cocktail had a half-life of ~100 hours in hamsters. Lastly, they measured antibody binding kinetics using a plasmon resonance (SPR) device.
Study findings
The key study finding was that IMM-BCP-01, a combination of IMM20253, IMM20190, and IMM20184 antibodies, bound to conserved non-overlapping epitopes of SARS-CoV-2 S trimer, dissociating it into S monomers and cleaving it into S1 and S2 subunits; finally, leading to complete reorganization of S.
Structural analysis of the S trimer complexed with the bound fragment, antigen-binding (Fabs) of IMM20184, IMM20190, or IMM20253 identified binding patterns of the IMM-BCP-01 antibodies. A three-dimensional (3D) reconstruction of cryo-EM micrographs of IMM20184 Fabs bound to S trimer revealed a 3:1 (Fab: Trimer) complex at ~10 angstroms (Å) resolution with a decreased density in the S core, indicating the S protein rearrangements.
Mutagenesis identified that IMM20184 bound to a highly conserved region in the core receptor-binding domain (RBD), a site close to the previously reported epitopes of CR3022 and COVA1-16 antibodies. The IMM20184 epitope included residues N370, K378, F374, and SP383-384 that were completely conserved among all current and previous SARS-CoV-2 VOCs, including Omicron BA.1 and BA.2 variants. These findings were further successfully validated using cryo-EM analysis.
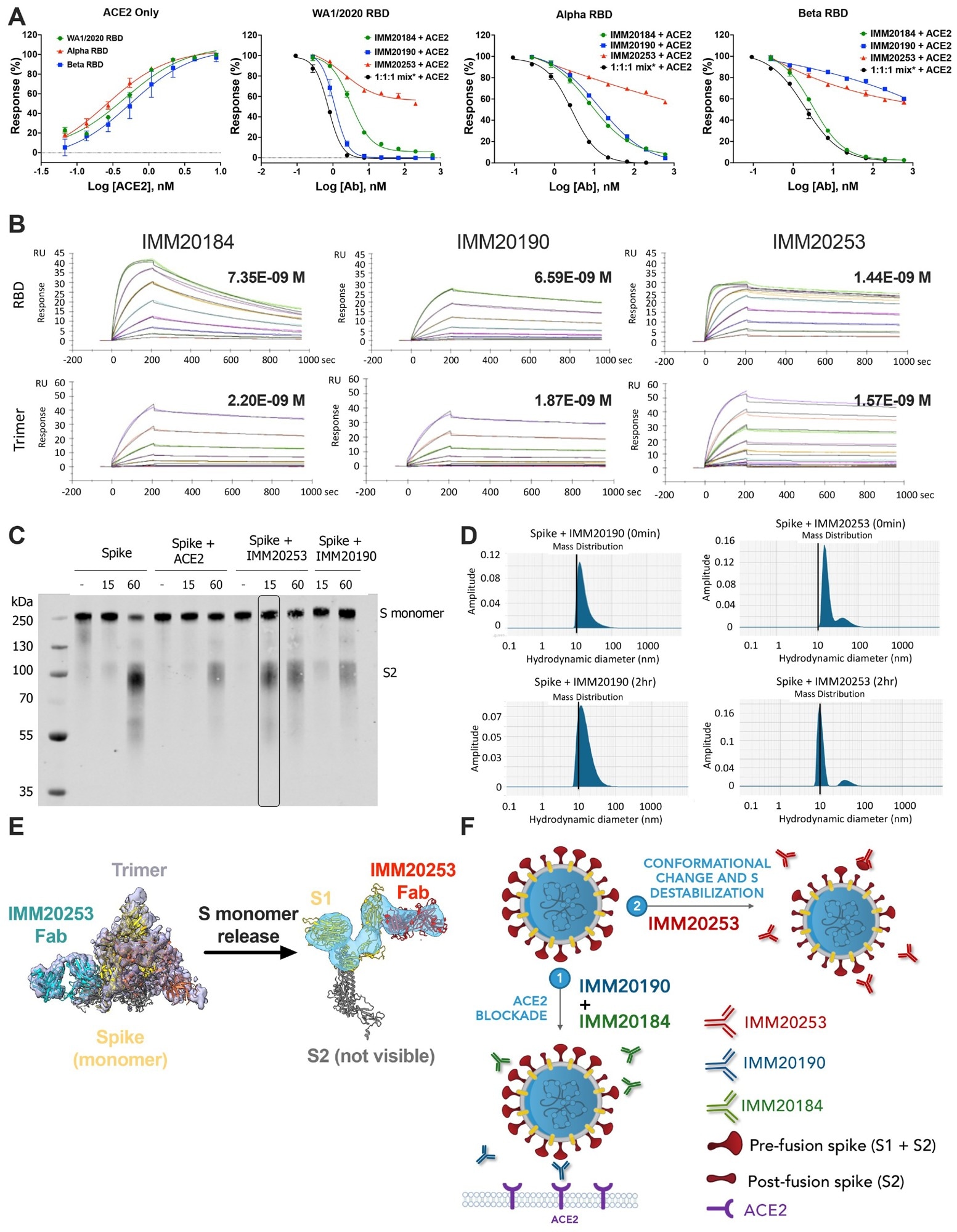 IMM20253 antibody inhibits virus in non-ACE2 dependent manner and facilitates the release of S1 protein. (A) Inhibition of RBD binding to its cellular receptor ACE2 in the presence of IMM20184/190/253. ELISA-based receptor competition assay. Denoted points are means of three replicates. Error bars denote SD. (B) Antibody binding kinetics of IMM20184, IMM20190 and IMM20253 antibodies to soluble RBD and Trimer (WA1/2020 variant) measured using Surface Plasmon Resonance (SPR). Denoted values are KD. (C). Western blot analysis of Trimer digested with protease K after 0, 15 and 60 min in the presence of either human ACE2, IMM20253 or IMM20190. Anti-S2 staining reveals S monomer (S1+S2) and S2 protein. (D) Dynamic light scattering (DLS) analysis of Trimer complex with IMM20253 or IMM20190 immediately or after 2 hours incubation measures a hydrodynamic diameter of each complex in nm. (E) IMM20253 Fab binding to Trimer triggers complex disruption and release of S monomers. (F) Schematic of mechanism of action of the IMM-BCP-01 cocktail.
IMM20253 antibody inhibits virus in non-ACE2 dependent manner and facilitates the release of S1 protein. (A) Inhibition of RBD binding to its cellular receptor ACE2 in the presence of IMM20184/190/253. ELISA-based receptor competition assay. Denoted points are means of three replicates. Error bars denote SD. (B) Antibody binding kinetics of IMM20184, IMM20190 and IMM20253 antibodies to soluble RBD and Trimer (WA1/2020 variant) measured using Surface Plasmon Resonance (SPR). Denoted values are KD. (C). Western blot analysis of Trimer digested with protease K after 0, 15 and 60 min in the presence of either human ACE2, IMM20253 or IMM20190. Anti-S2 staining reveals S monomer (S1+S2) and S2 protein. (D) Dynamic light scattering (DLS) analysis of Trimer complex with IMM20253 or IMM20190 immediately or after 2 hours incubation measures a hydrodynamic diameter of each complex in nm. (E) IMM20253 Fab binding to Trimer triggers complex disruption and release of S monomers. (F) Schematic of mechanism of action of the IMM-BCP-01 cocktail.
It is noteworthy that the IMM20253 epitope is present in all SARS-related coronaviruses (CoVs); therefore, it might remain conserved across emerging SARS-related human CoVs. Also, IMM20253 triggered a conformational change of S protein into a post-fusion form to prevent angiotensin-converting enzyme 2 (ACE2)-dependent host receptor binding. It bound a conserved epitope, with K356 and R466 residues, not a common target of the human immune system for generating neutralizing antibodies.
The three-antibody cocktail consistently showed robust antiviral potency in vivo and in vitro, neutralized all VOCs tested, including Omicron BA.1 and Omicron BA.2, and induced a potent antiviral effector response. Notably, all three antibodies elicited viral neutralization through different mechanisms. The neutralization potency exhibited by IMM-BCP-01 across pseudo- and live virus assays translated to a higher in vivo efficacy in animal models. Notably, IMM-BCP-01 was particularly highly effective against the Beta and Omicron VOCs.
Further, the authors demonstrated that the IMM-BCP-01 cocktail accentuated fraction, crystallizable (Fc)-mediated SARS-CoV-2 clearance mechanisms in vivo more than any of the three individual antibodies alone. Accordingly, it robustly neutralized SARS-CoV-2 VOCs utilizing multiple mechanisms. More importantly, the dose-response of the IMM-BCP-01 cocktail was not limited to the WA1/2020 SARS-CoV-2 isolate. It was also efficacious in clearing Alpha, Beta, and Omicron VOCs within the three to nine mg/kg dose range.
Conclusions
The study demonstrated that a rare cocktail of patient-derived IgGs, the IMM-BCP-01 cocktail, specific to conserved non-overlapping epitopes on S protein potently activated Fc-mediated antiviral effector response and demonstrated antiviral effects in vivo. In addition, it remained resistant to mutational drift.
Further, the pre-clinical study data confirmed that IMM-BCP-01 was consistently effective across the spectrum of SARS-CoV-2 variants, including Alpha, Beta, Gamma, Delta, Epsilon, Kappa, Lambda, Mu, Zeta, and Omicron BA.1 and BA.2 variants in vitro. Overall, the IMM-BCP-01 antibody cocktail could be a promising prophylactic and therapeutic against SARS-CoV-2; therefore, the authors emphasized moving it quickly into clinical trials.
Journal reference:
- IMM-BCP-01, a patient-derived anti-SARS-CoV-2 antibody cocktail, is active across variants of concern including Omicron BA.1 and BA.2, Pavel A. Nikitin, Jillian M. Dimuzio, John P. Dowling, Nirja B. Patel, Jamie L. Bingaman-Steele, Baron C. Heimbach, Noeleya Henriquez, Chris Nicolescu, Antonio Polley, Eden L. Sikorski, Raymond J. Howanski, Mitchell Nath, Halley Shukla, Suzanne M. Scheaffer, James P. Finn, Li-Fang Liang, Todd Smith, Nadia Storm, Lindsay G. A. Mckay, Rebecca I. Johnson, Lauren E. Malsick, Anna N. Honko, Anthony Griffiths, Michael S. Diamond, Purnanand Sarma, Dennis H. Geising, Michael J. Morin, Matthew K. Robinson, Science Immunology 2022, DOI: 10.1126/sciimmunol.abl9943, https://www.science.org/doi/10.1126/sciimmunol.abl9943