An article under review in the Scientific Reports journal and currently available on the Research Square* preprint server reports on chicoric acid (CA) as a potential inhibitor of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
There is still a critical medical need for novel, targeted medications that can combat SARS-CoV-2 infections. Establishing such antiviral agents, in most instances, demands the knowledge of the function and structure of possible viral targets and the identification and profiling of small-molecule modulatory attachment sites in these proteins that allow structure-based drug design (SBDD) techniques.
The SARS-CoV-2 nucleocapsid (N) protein is an important target for developing new antivirals since it is essential for the CoV genome transcription and packaging. The N protein is expressed most frequently in infected cells. Yet, it has not been utilized widely as a target for anti-SARS-CoV-2 drug development, in contrast to the envelope (E) and spike (S) structural proteins. Indeed, the structural data on the N protein's ligand-binding is limited.
About the study
In the present study, the researchers from the Brazilian Center for Research in Energy and Materials and the State University of Campinas utilized a new fluorescence polarization-based high-throughput screening (HTS) assay to uncover small compounds that interfere with the N protein's ability to bind to a specific RNA, i.e., RNA1, obtained from the SARS-CoV-2 genome packing signal (PS).
To develop an assay for recognizing the possible inhibitors of the N protein RNA-binding capability, the team searched for an N protein target RNA employing the SARS-CoV-2 PS sequence as the candidate. A fluorescence polarization (FP) test using different 5'-fluorescein isothiocyanate (FITC)-labeled RNAs as targets tracked the N protein's RNA-binding activities.
The scientists used an FP assay to run an HTS experiment examining a library of about 3200 authorized medicines and bioactive compounds. Concentration-response studies were conducted using the shortlisted molecules to verify the hit candidates' efficacy as inhibitors of the N protein-RNA1 contact.
1H-saturation transfer difference nuclear magnetic resonance (1H-STD NMR) and isothermal titration calorimetry (ITC) were used to further examine the characteristics of CA as a ligand for the N protein. The authors established the crystal structure of the N protein's C-terminal domain (CTD) coupled with CA to shed light on how CA binds to the N protein. They sought to see if CA may prevent SARS-CoV-2 infection in vitro after validating CA as an N protein-ligand with probable consequences on its RNA attachment activity.
Results
In the assessment of bioactive small molecules immensely polar compounds stood out, particularly polyphenols like chebulinic acid (CI), ellagitannins, punicalin (PL), and punicalagin (PG), as well as tartaric acid diesters, such as CA, and polysulphonated naphthylureas like suramin (SUR). The latter substances interfered with the contact of an RNA probe generated from the SARS-CoV-2 PS sequence, RNA1, and the full-length SARS-CoV-2 N protein at the submicromolar level.
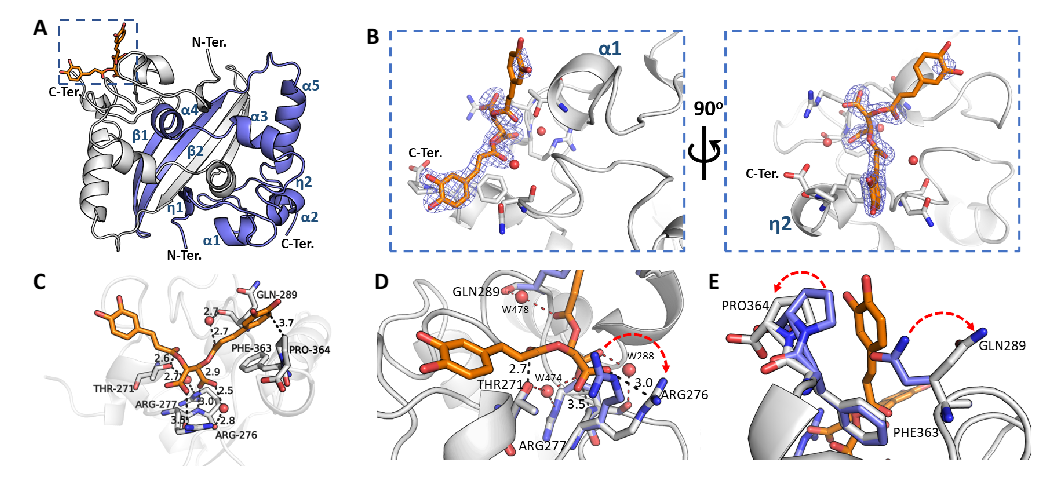
The crystal structure of SARS-CoV-2 N protein CTD binding chicoric acid (CA) reveals a network of polar contacts and structural readjustments to accommodate the symmetric ligand in the CA-N protein binding site. A) Cartoon representation of the N protein CTD dimer crystal structure depicting its secondary structure elements (in blue), including two 310 (η) helices, five α-helices and two antiparallel β-strands and the CA binding site (inset highlighted by the blue dashed square). B) Detailed CA-binding site from the inset of panel A. The CA molecule is represented as sticks (orange) with its electron-density map in blue. CA binds to a shallow pocket formed between α-helices 1-2 and η-helix 2, close to the C-terminus (C-Ter). C) CA atomic interactions with the N protein residues. The CA carboxyl groups are at ideal distances to engage electrostatic interactions and hydrogen bonds with Arg276 side chain (NH1 atom), Arg277 main chain amine and a structural water molecule (W288) stabilized by Arg276 NH2. Thr271 and Gln289 can further position hydrogen bond donors (Thr271O�� and a structural water molecule, W478, stabilized by the Gln289 carbonyl) to engage a symmetric interaction with the carbonyl groups from both caffeoyl units of CA. One of the catechol motifs of CA is well accommodated near the C-terminal Pro364, showing a well-defined electron density (vide panel B). D, E) Superposition of the CA-binding site in the native N protein CTD (blue sticks, PDB ID 7UXX) and CA-N protein CTD complex (grey sticks (PDB ID 7UXZ)) highlighting structural readjustments induced by CA binding (highlighted by red arrows). Figures were generated with Pymol (Schroedinger Inc.). Polar contacts are indicated by dashed lines with the measured distances in Angstroms. Oxygen atoms are shown in red, nitrogen atoms in blue. Water molecules are represented as red spheres.
CI, PL, PG, and SUR were described priorly to display different biological characteristics, including antiviral action. However, CA was highlighted as a novel class of N protein modulators and one of the most effective hit compounds discovered in the present HTS trials.
The findings demonstrated that CA was an affinity ligand for the N protein that attaches to the CTD and expels the RNA from the N protein at micromolar levels. The team found that CA suppresses SARS-CoV-2 multiplication in cell culture at micromolar concentrations, which was compatible with the dissociation constant (KD) values for the dissociation of the N protein and RNA1 complex.
Conclusions
According to the study's authors, the present research was the first characterization of a non-endogenous ligand for the SARS-CoV-2 N protein and the initial account of this modulatory ligand binding location on the CoV N protein.
In the current study, the scientists described a novel fluorescence-based HTS test that enables the discovery of small compounds that obstruct the SARS-CoV-2 N protein's ability to bind RNA. They used a series of biophysical studies to characterize the top hits. The researchers solved the crystal structure of the non-endogenous ligand binding the N protein CTD for the first time, illuminating a novel modulatory region in the SARS-CoV-2 N protein. Notably, the CA-binding region was conserved in SARS-CoV and partly conserved in the N proteins of the Middle East respiratory syndrome (MERS).
The current data provide the structural foundation for the rational design and establishment of new antiviral medicines addressing the SARS-CoV-2 N protein, a relevant and still unstudied target of CoVs, despite the necessity for further refinement of CA as an antiviral agent.

 *Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor; Gustavo Mercaldi, Eduardo Bezerra, Fernanda Aparecida Batista, Celisa Tonoli, Adriana Soprano, Jacqueline Shimizu, Alice Nagai, Jaqueline da Silva, Helder Ribeiro-Filho, Jessica Faria, Marcos da Cunha, Ana Zeri, Andrey Fabricio Nascimento, Jose Luiz Proenca-Modena, Marcio Bajgelman, Silvana Rocco, Paulo Lopes-de-Oliveira, Artur Cordeiro, Marjorie Bruder, Rafael Elias Marques, Mauricio Sforca, Kleber Franchini, Celso Benedetti, Ana Carolina Figueira, Daniela Trivella. Research square preprint. DOI: https://doi.org/10.21203/rs.3.rs-1720953/v1, https://www.researchsquare.com/article/rs-1720953/v1