This AA stretch is also most prone to mutations; subsequently, in Alpha and Delta SARS-CoV-2 variants of concern (VOCs), several mutations are harbored between Pro681 to histidine (His) and arginine (Arg), respectively. In all Omicron sub-lineages, including BA.1, BA.2, and BA.5, there are mutations at P681H and N679K. All these mutations have S proteolytic cleavage enhancing effect. It is an analytical challenge to study this peptide stretch in SARS-CoV-2 S due to its biosynthetic complexity and the lack of a peptide consensus sequence. Moreover, there is a lack of data on glycan abundance, their attachment site(s), and their structural influence on proteolysis. Furthermore, studies have barely investigated the role of SARS-CoV-2 VOC mutations on O-GalNAc glycosylation.
There’s a family of 20 GalNAc transferase, GalNAc-T1 to T20 isoenzymes with varying substrates. Identifying glycosylation sites of each GalNAc-T could provide insights into O-glycan biology. In a previous study by Ten Hagen et al., they found seven isoenzymes that could introduce GalNAc into SARS-CoV-2 S.
About the study
In the present study, researchers used a chemical engineering tactic termed “bump-and-hole (BH) engineering” to study individual GalNAc-T isoenzymes in the living cell. Further, they insisted that improving fragmentation-based sequencing of glycopeptides could enable enhanced mass spectrometry(MS)-based analysis.
First, the researchers incubated recombinant S of SARS-CoV-2 wild-type (WT) strain with P681R (or P681H) constructs produced in human Expi293-F cells with either WT- or BH-versions of GalNAc-T1 or T2. Then, they proceeded to bump these constructs with nucleotide-sugar uridine diphosphate (UDP)-GalN6yne. They tagged glycosylated versions by copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) for visualization via the Streptavidin blot. This treatment introduced GalN6yne in an isoenzyme-specific fashion while endowing glycopeptides with an additional positive charge that facilitated MS-analysis.
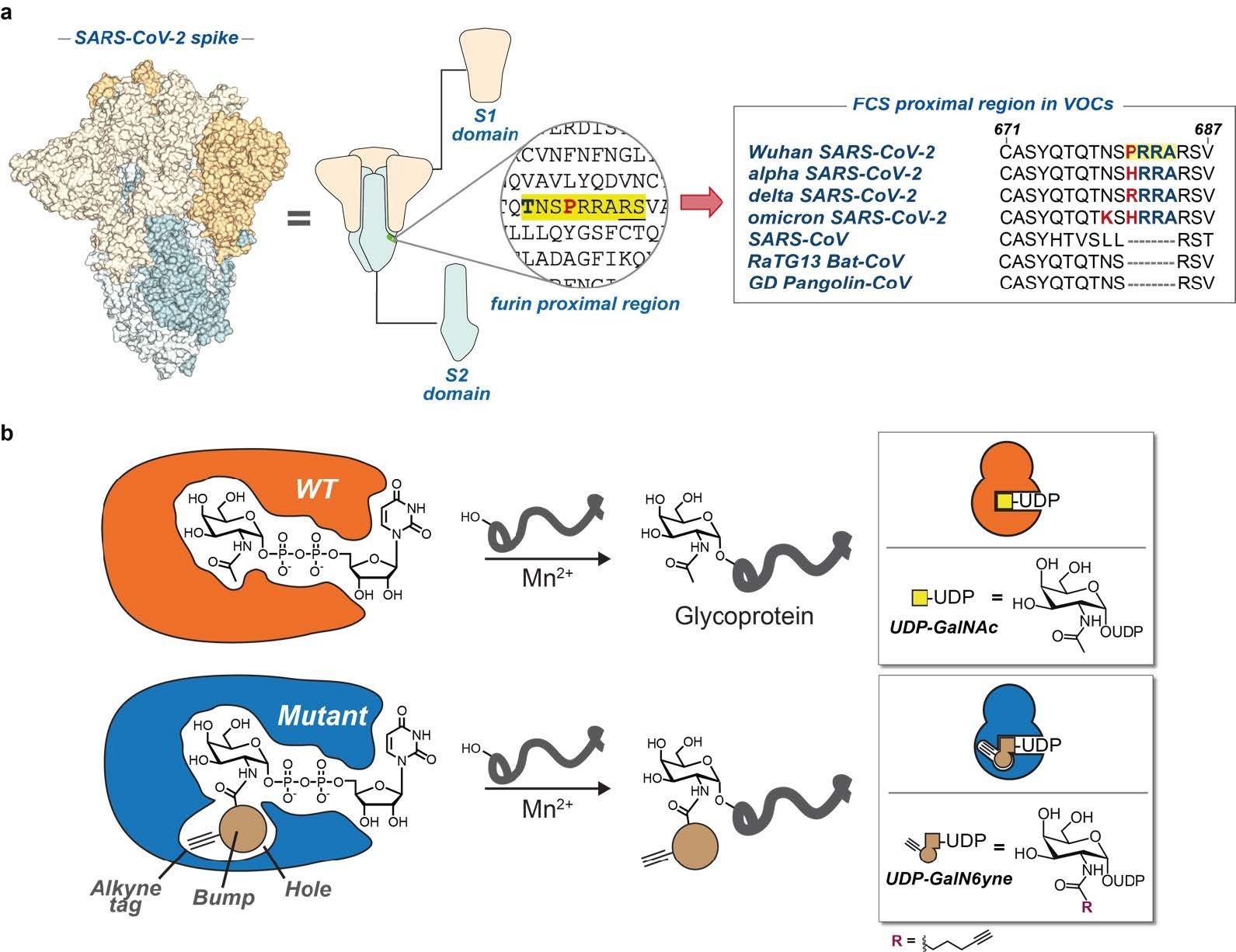
a) SARS-CoV-2 spike model and cartoon. Furin cleavage site and a peptide alignment for COVID- 19 variants of concern and related coronaviruses. Highlighted in yellow is the polybasic motif of SARS-CoV- 2. Bold and red are amino acid changes in positions 679 and 681. b) Bump-and-hole engineering allows for GalNAc-T isoenzyme-specific tagging of glycosylation substrates using the clickable substrate UDPGalN6yne. FCS = furin cleavage site; VOCs = variants of concern.
Then, the team subjected FL-S and S1/S2 subunits to in-gel digestion and analyzed them by tandem MS (ETD). They used high-intensity collision-induced dissociation (HCD) to obtain naked peptide sequences and glycan compositions and then used the I-tagged-GalN6yne diagnostic ion to trigger ETD fragmentation of the peptide backbone. The researchers also validated the study results in vitro. To this end, they used glycosylation of recombinantly expressed S proteins with soluble versions of BH-GalNAc-T1 and BH-GalNAc-T2 and CuAAC with I-tagged-azide and MS glycoproteomic analysis.
The team used a panel of synthetic peptides to study the effect of S protein mutations on GalNAc-T1-mediated glycosylation. These peptides had SARS-CoV-2 VOC-related mutations at the major hotspots: Gln675, Gln677, asparagine (Asn)679, and Pro681.
Glycosylation modulates S proteolytic processing depending on the distance to the cleavage site and glycan composition. So, the team directly probed whether O-GalNAc glycans on Thr678 modulated S cleavage by furin using synthetic Förster Resonance Energy Transfer (FRET)- active substrate peptides. Peptides spanning Gln residues 672 to 689 contained N-terminal 2-aminobenzoyl and C-terminal 3-nitro-Tyr as fluorescence donor and quencher moieties, respectively. An increase in fluorescence intensity indicated furin-mediated S cleavage. First, they compared non-glycosylated substrates corresponding to either WT (FRET-1) and P681H mutant spike (FRET-2). In addition, they subjected recombinant TMPRSS2 to glycopeptide FRET substrates FRET-1, FRET-3, and FRET-7 to FRET-9.
Study findings
Computational and manual validation revealed that Thr678 carried I-tagged-modified GalN6yne in both FL-S and S1 samples only in cells expressing BH-T1. Both BH-T1 and BH-T2 glycosylated threonine (Thr)323 had not been associated with any GalNAc-T isoenzyme. FRET analysis revealed that elaboration of O--GalNAc glycans on Thr678 of SARS-CoV-2 S glycoprotein, especially with negatively charged modifications, severely hampered the furin activity. The O-glycans in the lung epithelial cells expressing GalNAc-T1 suggested that glycosylation is a physiologically significant modification that restricts the maturation of S protein during the course of evolution.
According to the authors, disruption of O-GalNAc glycosylation was a major driving force behind the evolution of SARS-CoV-2 VOCs. Within the evolutionary trajectory from the SARS-CoV-2 Alpha to Delta and Omicron VOCs, notable changes in the AA sequences preceding the FCS indicated that proteolytic cleavage gradually enhanced with the increase in VOC infectivity. The analogous mutation found on the more transmissible Delta VOC (P681R) has also been linked to a rise in furin cleavage, suggesting an evolutionary trajectory that suppresses O-glycosylation before increasing intrinsic furin recognition.
In contrast to P681H detected in early variants such as Alpha, the N679K mutation in Omicron did not substantially impact glycosylation but led to enhanced furin cleavage of synthetic peptides. These FCS-proximal mutations acted synergistically and evolved to suppress glycosylation and enhance furin cleavage.
Conclusions
The current study established that O-glycosylation is a key determinant of SARS-CoV-2 S cleavage and maturation. It might also be a remnant of ancestral SARS-CoV-2 strains lost through viral evolution.
The researchers evidenced that GalNAc-T1 primed glycosylation at Thr678 in the living cells. The glycosylation site at Thr678 of the S (not close to the FCS) explained why a single GalNAc residue was insufficient to modulate furin activity and only slightly impacted TMPRSS2 activity. On the contrary, elaboration and sialyation abrogated furin activity by up to 65%. Similarly, O-glycosylation negatively impacted S cleavage by TMPRSS2, with core 1 (Galβ1-3GalNAc-) disaccharide-containing O-glycans having the most effect. Overall, the study findings emphasized the need for more advanced glycan tracing techniques to study SARS-CoV-2 evolution.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
O-Linked Sialoglycans Modulate the Proteolysis of SARS-CoV-2 Spike and Contribute to the Mutational Trajectory in Variants of Concern, Edgar Gonzalez-Rodriguez, Mia Zol-Hanlon, Ganka Bineva-Todd, Andrea Marchesi, Mark Skehel, Keira Mahoney, Chloe Roustan, Annabel Borg, Lucia Di Vagno, Svend Kjaer, Antoni G. Wrobel, Donald J. Benton, Philipp Nawrath, Sabine L. Flitsch, Dhira Joshi, Andres Manuel Gonzalez-Ramirez, Katalin A. Wilkinson, Robert J Wilkinson, Ramon Hurtado-Guerrero, Stacy A Malaker, Benjamin Schumann, bioRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.09.15.508093, https://www.biorxiv.org/content/10.1101/2022.09.15.508093v2
- Peer reviewed and published scientific report.
Gonzalez-Rodriguez, Edgar, Mia Zol-Hanlon, Ganka Bineva-Todd, Andrea Marchesi, Mark Skehel, Keira E. Mahoney, Chloë Roustan, et al. 2023. “O-Linked Sialoglycans Modulate the Proteolysis of SARS-CoV-2 Spike and Likely Contribute to the Mutational Trajectory in Variants of Concern.” ACS Central Science 9 (3): 393–404. https://doi.org/10.1021/acscentsci.2c01349. https://pubs.acs.org/doi/10.1021/acscentsci.2c01349.