Several studies have reported that coronavirus disease 2019 (COVID-19) vaccine boosters enhance immune responses to ancestral SARS-CoV-2 and emergent variants of concern. Besides, new vaccines based on circulating SARS-CoV-2 variants are being developed to augment antibody responses.
Moreover, recent evidence suggests that a booster dose based on the Beta variant results in higher titers of neutralizing antibodies (nAbs) against Beta and Omicron variants than wildtype-based booster or bivalent vaccines encoding wildtype and Omicron spike proteins. Nonetheless, it remains unclear whether booster doses induce GC reactions.
 Study: SARS-CoV-2 Omicron boosting induces de novo B cell response in humans. Image Credit: NIAID
Study: SARS-CoV-2 Omicron boosting induces de novo B cell response in humans. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The study and findings
In the present study, researchers evaluated the immune responses of SARS-CoV-2 infection-naïve healthy, vaccinated adults. Participants completed primary vaccination with Pfizer’s BNT162b2 or Moderna’s mRNA-1273 vaccine. They were boosted with a single dose of mRNA-1273 or the bivalent mRNA-1273.213 (based on the spike proteins of Beta and Delta variants). Forty-six individuals were recruited; seven were boosted with the mRNA-1273, while 31 received the variant-specific vaccine.
Enzyme-linked immunosorbent spot (ELISpot) assay was used to quantify circulating spike-specific plasmablasts. The authors detected spike-specific IgA- and IgG-producing plasmablasts in all mRNA-1273 recipients a week after boosting. Similarly, they observed IgG-producing plasmablasts against the spike protein of ancestral strain, Beta, and Delta variants. In addition, fine needle aspirates (FNAs) and bone marrow aspirates were collected from some individuals post-boost.
FNA samples were stained with fluorescently-labeled spike protein probes from ancestral strain, Beta, Delta, and Omicron (BA.1) variants. Spike-binding GC B lymphocytes and follicular T helper cells were detected in all FNA samples from all participants after two weeks. Spike-specific memory B cells (MBCs) were present in all participants before booster vaccination, which were sustained at similar frequencies post-boost.
Three participants from mRNA-1273 and mRNA-1273.213 cohorts were selected to characterize the MBC repertoire. Clonally distinct antigen-specific monoclonal antibodies (mAbs) were generated from six participants and assessed for spike-binding using a multiplex bead binding assay. The authors noted that 94% and 92% of MBC-derived mAbs from mRNA-1273 and mRNA-1273.213 cohorts recognized spikes from ancestral strain, Beta, Delta, and BA.1 variants.
Single-cell RNA sequencing was performed on plasmablasts and FNA samples from these six individuals. Most spike-specific plasmablasts identified post-boost were clonally related to GC B cells, MBCs, and plasma cells elicited by primary vaccination. There were multiple spike-specific plasmablast clones after administration of second and booster doses. Representatives of these clones in booster responses showed significantly higher somatic hypermutation (SHM) frequencies.
Spike-specific plasmablast clones were identified in GC responses in all six individuals. SHMs in spike-specific MBCs were significantly higher post-boost than after primary vaccination. Although both booster vaccines induced robust GC response and MBC maturation, variant-specific antibodies (against Beta and Delta spikes) were not isolated, implying that the primary vaccination series imprinted the B cell response.
Further, eight individuals were recruited to assess whether an antigenically more divergent vaccine booster could result in immune responses against novel epitopes. These double-vaccinated subjects received the mRNA-1273.529 vaccine (booster) encoding the BA.1 spike. MBCs were sorted from peripheral blood samples of participants 17 weeks post-boost. Almost all mAbs (99%) were cross-reactive and bound to spikes from ancestral strain, Beta, Delta, and BA.1 variants.
Next, the neutralizing capacity of the mAbs from the three cohorts was examined in a high-throughput assay. The researchers detected 131 mAbs that neutralized infection by 80% at least; these mAbs were tested for neutralization against a panel of authentic SARS-CoV-2 particles. Most mAbs inhibited infection by 90% against ancestral strain, Beta, and Delta variants; however, there were relatively fewer mAbs in each cohort that were effective against BA.1 and BA.5 variants.
Lastly, the research team isolated mAbs that (specifically) recognized BA.1 spike that did not bind to the ancestral spike. Seventy-eight mAbs were isolated via this approach; of these, there were 57 mAbs that were still cross-reactive (to ancestral spike). One mAb recognized BA.1 spike, 12 recognized BA.1 and Beta/Delta spike, and eight mAbs did not bind to any antigen above background. None of these 13 mAbs neutralized the ancestral strain; seven (54%) mAbs neutralized BA.1, one mAb neutralized BA.5, while none of the mAbs neutralized Beta or Delta variants.
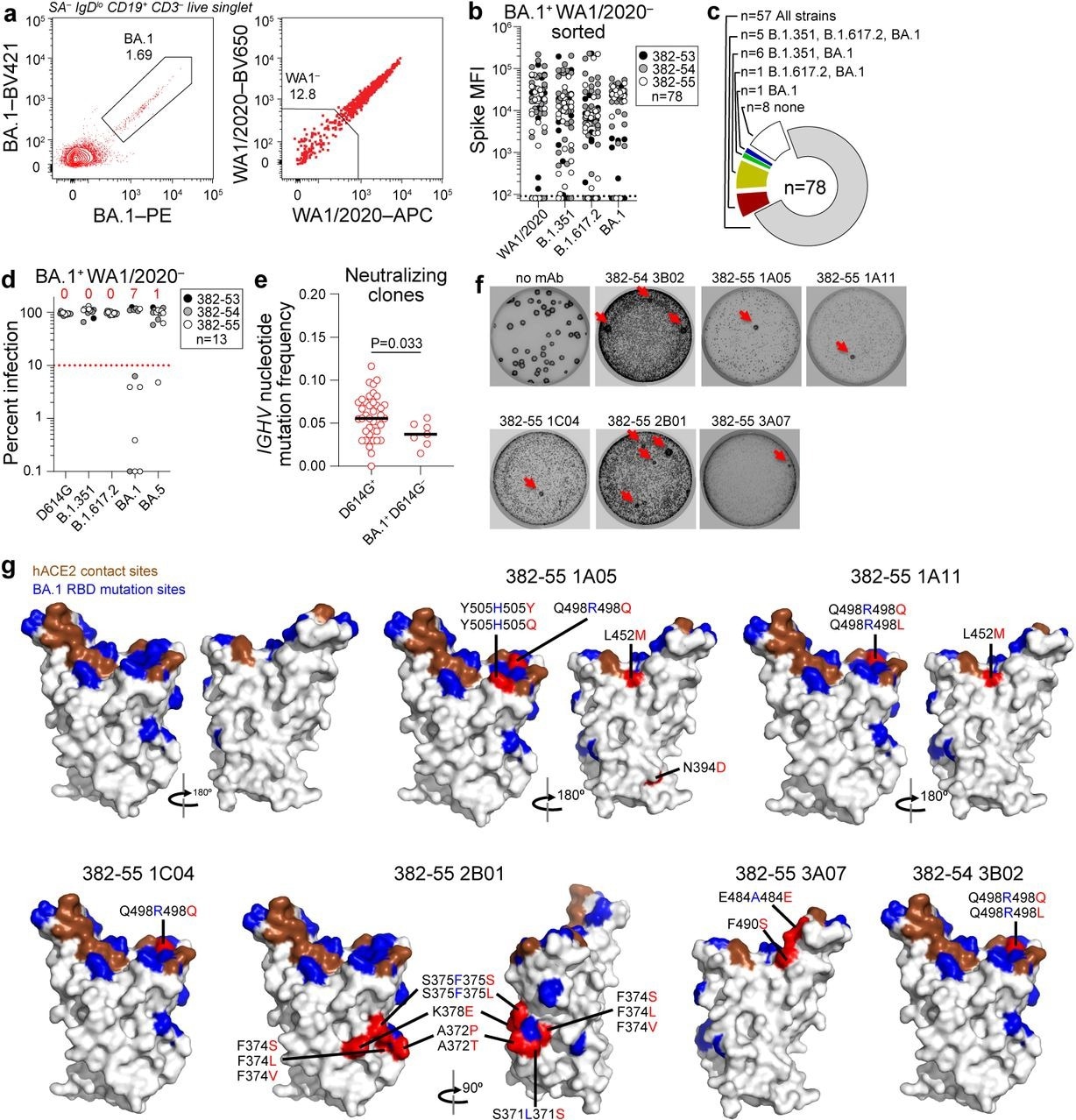
Characterization of BA.1-specific mAbs. (a) Gating strategy for sorting BA.1+ WA1/2020− MBC from 17 weeks post-boost PBMC. (b) Binding of mAbs from BA.1+ WA1/2020− sorted MBCs to indicated strains of SARS-CoV-2 S measured by multiplex bead binding array. (c) Summary of mAb binding. (d) Neutralizing activity of BA.1+ WA1/2020− binding mAbs against indicated strains of authentic SARS-CoV-2 virus. Numbers above each virus are of mAbs below the 90% infection reduction threshold. (e) IGHV mutation frequencies of clones related to mAbs from participants 382-54 and 382-55 that neutralized D164G (left) and BA.1 but not D614G (right). Black lines indicate medians. Each symbol represents a sequence; n = 39 for D614G+ and n = 7 for BA.1+ D614G−. (f) Plaque assays on Vero E6 cells with indicated mAb in the overlay to isolate escape mutants (red arrows). Images are representative of three experiments per mAb. (g) Structure of RBD with hACE2 footprint highlighted in brown, BA.1 mutations highlighted in blue, and amino acids whose substitution confers resistance to indicated mAbs in plaque assays highlighted in red.
Conclusions
The study illustrated that boosting with ancestral spike-based or bivalent (Beta/Delta) vaccine-induced potent spike-specific GC responses in axillary lymph nodes of all participants. Spike-specific GC B cells and plasmablasts were predominantly derived from pre-existing clonal lineages. Notably, the researchers showed that boosting with a monovalent, antigenically-distant Omicron-based vaccine (mRNA-1273.529) induced de novo B cell responses, targeting the novel epitopes of the Omicron spike.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Alsoussi WB, Malladi SK, Zhou JQ, et al. SARS-CoV-2 Omicron boosting induces de novo B cell response, DOI: 10.1101/2022.09.22.509040, https://www.biorxiv.org/content/10.1101/2022.09.22.509040v1
- Peer reviewed and published scientific report.
Alsoussi, Wafaa B., Sameer Kumar Malladi, Julian Q. Zhou, Zhuoming Liu, Baoling Ying, Wooseob Kim, Aaron J. Schmitz, et al. 2023. “SARS-CoV-2 Omicron Boosting Induces de Novo B Cell Response in Humans.” Nature, April, 1–3. https://doi.org/10.1038/s41586-023-06025-4. https://www.nature.com/articles/s41586-023-06025-4.