Background
The World Health Organization (WHO) has classified the SARS-CoV-2 Alpha, Beta, Gamma, Delta, and Omicron variants as variants of concern because they carry mutations in their spike protein that increase their transmissibility and ability to evade vaccine-induced immunity, as compared to the ancestral Wuhan strain.
The N501Y mutation in the receptor binding domain (RBD), which is reported to increase transmissibility by 40–70%, is shared by the Alpha, Beta, Gamma, and Omicron variants. Additionally, the Beta, Gamma, and Omicron variants also carry substitution mutations in the E484 and K417 positions, while the D614G mutation is present in all five variants of concern. A modified vaccine antigen containing substitution mutations shared by the variants of concern could elicit broadly neutralizing antibodies against the circulating variants.
About the study
The present study used the HexaPro (S-6P) S-trimer and an Alum/cytosine phosphoguanine (CpG) 7909 dual adjuvant system to create a modified spike trimer vaccine containing the four spike protein substitution mutations commonly found in the five variants of concern. Vero-E6 cells were used to propagate SARS-CoV-2 viruses of the ancestral strain and the variants of concern. The modified S-trimer was produced by synthesizing and cloning the codon-optimized gene into a mammalian expression vector.
The trimeric conformation and purity of the modified spike protein were tested using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and size-exclusion high-performance liquid chromatography (HPLC). The modified spike-trimer’s binding affinity to the human angiotensin-converting enzyme-2 (ACE-2) receptor and other kinetic properties were determined using biolayer interferometry assays.
The animal models used to test immunogenicity and antibody responses to viral challenge included BALB/c mice, rhesus macaques, and Golden Syrian hamsters. The animals were intramuscularly administered different vaccine doses, and the control and enzyme-linked immunosorbent assays (ELISA) and pseudovirus neutralization assays were used to determine the serum antibody titers. During the live virus neutralization experiments, plaque reduction neutralization assays were used to measure the macaque serum samples, while cytopathic effect neutralization assays were used to determine the neutralizing antibody titers in hamster serum samples.
Furthermore, peripheral blood mononuclear cells (PBMCs) or splenocytes harvested from the vaccinated animals 14 days after the second vaccine dose and analyzed through flow cytometry were used for intracellular cytokine staining assay and enzyme-linked immunospot (ELISPOT) assay to examine the immune responses elicited by the vaccines.
Additionally, quantitative reverse transcription polymerase chain reaction (qRT-PCR) was used to monitor SARS-CoV-2 ribonucleic acid (RNA) levels using swab samples and organ tissue from the animal models. Hamster and rhesus macaque lung tissues fixed in formalin and embedded in paraffin were used for hematoxylin and eosin staining and to study immunohistochemistry.
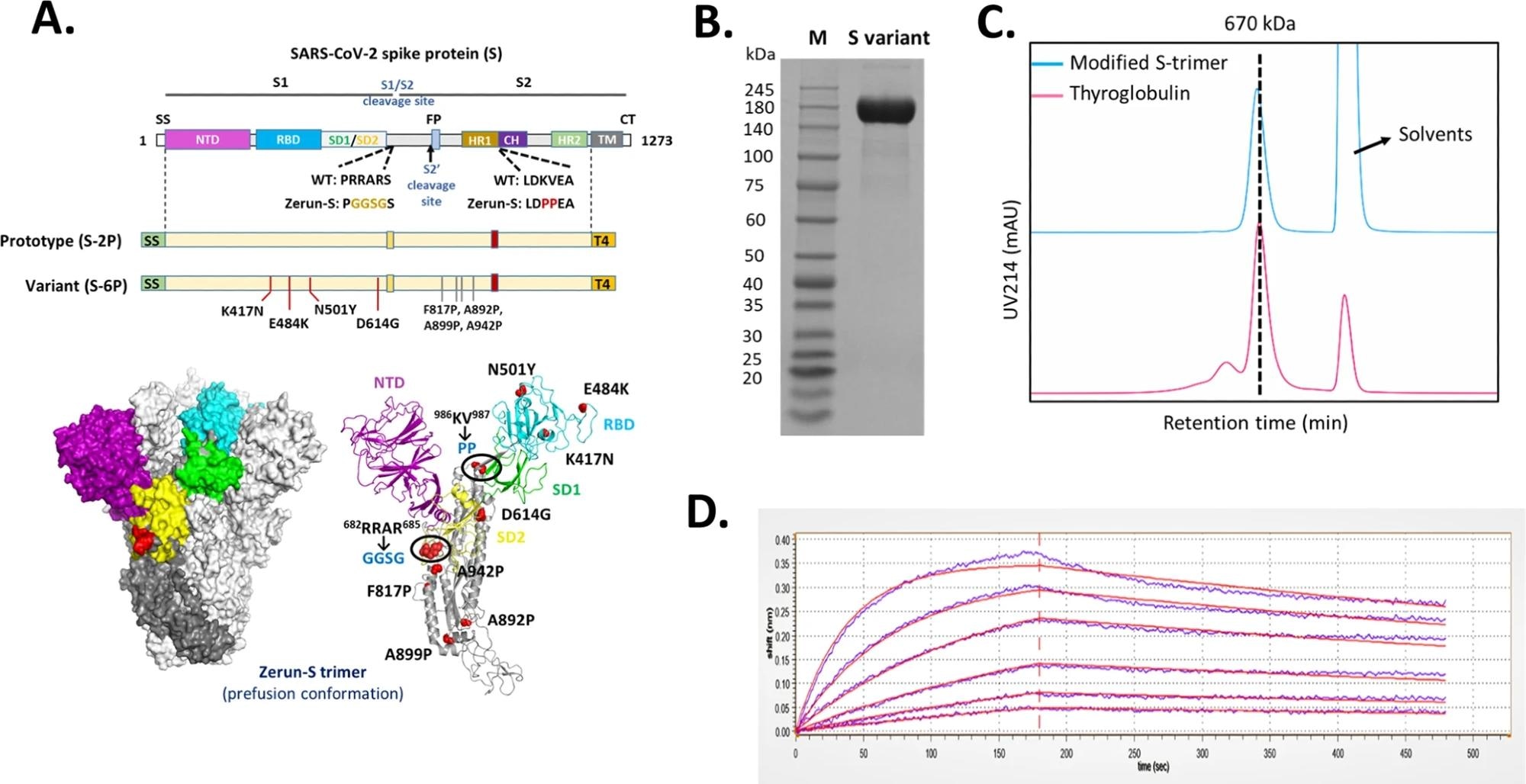 A Domain architecture of the SARS-CoV-2 S protein. SS signal sequence, NTD N-terminal domain, RBD receptor-binding domain, SD1 subdomain 1, SD2 subdomain 2, S1/S2 S1/S2 protease cleavage site, S2' S2' protease cleavage site, FP fusion peptide, HR1 heptad repeat 1, CH central helix, CD connector domain, HR2 heptad repeat 2, TM transmembrane domain, CT cytoplasmic tail. Prototype S-trimer (S-2P) contains two consecutive proline substitutions at residues 986 and 987, a “GGSG” substitution at the furin cleavage site, and a C-terminal T4 fibritin trimerization motif. Variant S-trimer (S-6P) contains additional four beneficial proline substitutions (F817P, A892P, A899P, and A942P), and four hot spot residues (K417N, E484K, N501Y, and D614G). The structure model of S-trimer was generated by the SWISS-MODEL using homology modeling techniques (http://swissmodel.expasy.org/), and the 3D structure figures were prepared using PyMOL (www.pymol.org). B SDS-PAGE analysis of purified variant S-trimer. Molecular weight standards are indicated at the left in kDa. C Size-Exclusion HPLC chromatogram of purified variant S-trimer (shown as cyan line) and a 670 kDa molecular weight standard (shown as purple line). D Binding profiles of variant S-trimer to human ACE2 measured by BLI in GatorPrime. The data are shown as blue lines, and the best fit of the data to a 1:1 binding model is shown in red.
A Domain architecture of the SARS-CoV-2 S protein. SS signal sequence, NTD N-terminal domain, RBD receptor-binding domain, SD1 subdomain 1, SD2 subdomain 2, S1/S2 S1/S2 protease cleavage site, S2' S2' protease cleavage site, FP fusion peptide, HR1 heptad repeat 1, CH central helix, CD connector domain, HR2 heptad repeat 2, TM transmembrane domain, CT cytoplasmic tail. Prototype S-trimer (S-2P) contains two consecutive proline substitutions at residues 986 and 987, a “GGSG” substitution at the furin cleavage site, and a C-terminal T4 fibritin trimerization motif. Variant S-trimer (S-6P) contains additional four beneficial proline substitutions (F817P, A892P, A899P, and A942P), and four hot spot residues (K417N, E484K, N501Y, and D614G). The structure model of S-trimer was generated by the SWISS-MODEL using homology modeling techniques (http://swissmodel.expasy.org/), and the 3D structure figures were prepared using PyMOL (www.pymol.org). B SDS-PAGE analysis of purified variant S-trimer. Molecular weight standards are indicated at the left in kDa. C Size-Exclusion HPLC chromatogram of purified variant S-trimer (shown as cyan line) and a 670 kDa molecular weight standard (shown as purple line). D Binding profiles of variant S-trimer to human ACE2 measured by BLI in GatorPrime. The data are shown as blue lines, and the best fit of the data to a 1:1 binding model is shown in red.
Results
The results reported broadly neutralizing antibody responses elicited by the novel variant vaccine against the ancestral Wuhan strain and the five variants of concern. In BALB/c mice models and rhesus macaques, the vaccine induced a significant CD4+ T helper cell response and stimulated the T helper type 1 cytokine profile.
During the viral challenge in Golder Syrian hamsters with the Beta and Delta variants, immunization with the recombinant vaccine decreased viral loads in the lung tissue and nasal turbinates and resulted in reduced lung inflammation and an increase in weight. The vaccine also decreased the viral loads in the lung and bronchus tissue and reduced viral shedding in the throat, blood, and anal swabs in rhesus macaques challenged with prototype SARS-CoV-2.
Two doses of the variant vaccine comprising the modified S-6P trimer and the alum/CpG 7909 adjuvant elicited potent cross-neutralizing antibodies and displayed post-challenge protection against the ancestral Wuhan strain and the Alpha, Beta, Gamma, Delta, and Omicron variants in mice, rhesus macaque, and hamster models.
Conclusions
Overall, the results from the neutralization assays and viral challenge tests indicated that the variant vaccine consisting of a modified S-6P trimer and a dual adjuvant of alum and CpG 7909 elicited potent cross-reactive neutralizing antibodies in hamsters, mice, and rhesus macaque models. The broad neutralizing ability against the ancestral Wuhan strain and the five major variants of concern make this variant vaccine a promising candidate for clinical trials.
Journal reference:
- Wang, Z., An, J., Liu, K., Yu, P., Fang, X., Li, J., Zhu, H., Zhu, Q., Huang, C., Zhang, C., Zhao, B., Bao, L., Song, Y., Cao, X., Hu, D., Jiang, Y., Shi, L., Zhou, L., Fan, J., & Guan, W. (2022). A potent, broadly protective vaccine against SARS-CoV-2 variants of concern. npj Vaccines, 7(1). https://doi.org/10.1038/s41541-022-00571-0, https://www.nature.com/articles/s41541-022-00571-0