The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by the highly contagious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has so far claimed more than 6.8 million lives worldwide.
 Study: A new generation Mpro inhibitor with potent activity against SARS-CoV-2 Omicron variants. Image Credit: Jezper / Shutterstock
Study: A new generation Mpro inhibitor with potent activity against SARS-CoV-2 Omicron variants. Image Credit: Jezper / Shutterstock
Background
Although several COVID-19 vaccines are commercially available, their efficacy has reduced due to the continual emergence of new COVID-19 variants, such as the Alpha, Beta, Delta, and the latest Omicron. Many SARS-CoV-2 variants can escape immune responses induced via vaccination or natural infection. Based on historical experience, there is a high risk of the long-term existence of SARS-CoV-2. Therefore, in addition to more effective COVID-19 vaccines, there is an urgent need for more antivirals against current and future SARS-CoV-2 variants.
SARS-CoV-2 belongs to the family Coronaviridae of genus β-coronavirus. This virus encodes 14 open reading frames (ORFs), which encode four structural proteins, nine accessory proteins, and two long polyproteins, pp1a and pp1ab. The polyproteins cleave into sixteen non-structural proteins (NSPs) using two cysteine proteases, namely, main protease (Mpro) and papain-like protease (PLpro).
Mpro, or 3CLpro, is a conserved gene of the virus and its variant. In addition, this protease is also present in other pathogenic coronaviruses, such as severe acute respiratory syndrome coronavirus-1 (SARS-CoV-1) and middle east respiratory syndrome coronavirus (MERS-CoV).
Mpro is linked to producing thirteen NSPs by cleaving pp1a and pp1ab. Mechanistically, Mpro identifies and cleaves amino acid sequences with specificity, particularly cleavage sites at sequences Leu-Gln. Importantly, no human proteases possess similar specificity.
Considering all attributes of Mpro, it can be a potential target for developing antivirals. Several small molecules, such as Remdesivir, Molnupiravir, and Paxlovid, have been approved as antiviral agents. Nirmatrelvir, Ensitrelvir, and Simnotrelvir are inhibitors of SARS-CoV-2 Mpro. However, these antivirals have some toxicity-related issues, suboptimal potency, and imperfect pharmacokinetic (PK) properties, such as poor oral bioavailability, poor oral bioavailability, and modest stability in human liver microsomes (HLM).
Notably, SARS-CoV-2 variants that contain Q192S/T/V, E166N/V, G143S, H172F/Q/Y, Q189E, M165T or A173V, have shown resistance to Nirmatrelvir treatment. Therefore, developing next-generation antivirals effective against SARS-CoV-2 variants is urgently needed.
The study and its findings
Primarily, scientists screened around 30,000 compounds from the in-house chemical library to obtain a new starting active compound to develop a drug targeting Mpro. They used fluorescence resonance energy transfer (FRET) assay for this purpose.
Four compounds were found to inhibit the enzymatic activity of Mpro, with 50% inhibition concentration (IC50) values less than 50 μM. Among these, Hit-1 was determined to be the most potent, with an IC50 value of 1.30 μM. A differential scanning fluorimetry (DSF) assay was used to validate the activity of Hit-1. This assay provided a thermal shift value of 8.45°C, which implies a direct binding between Hit-1 and Mpro.
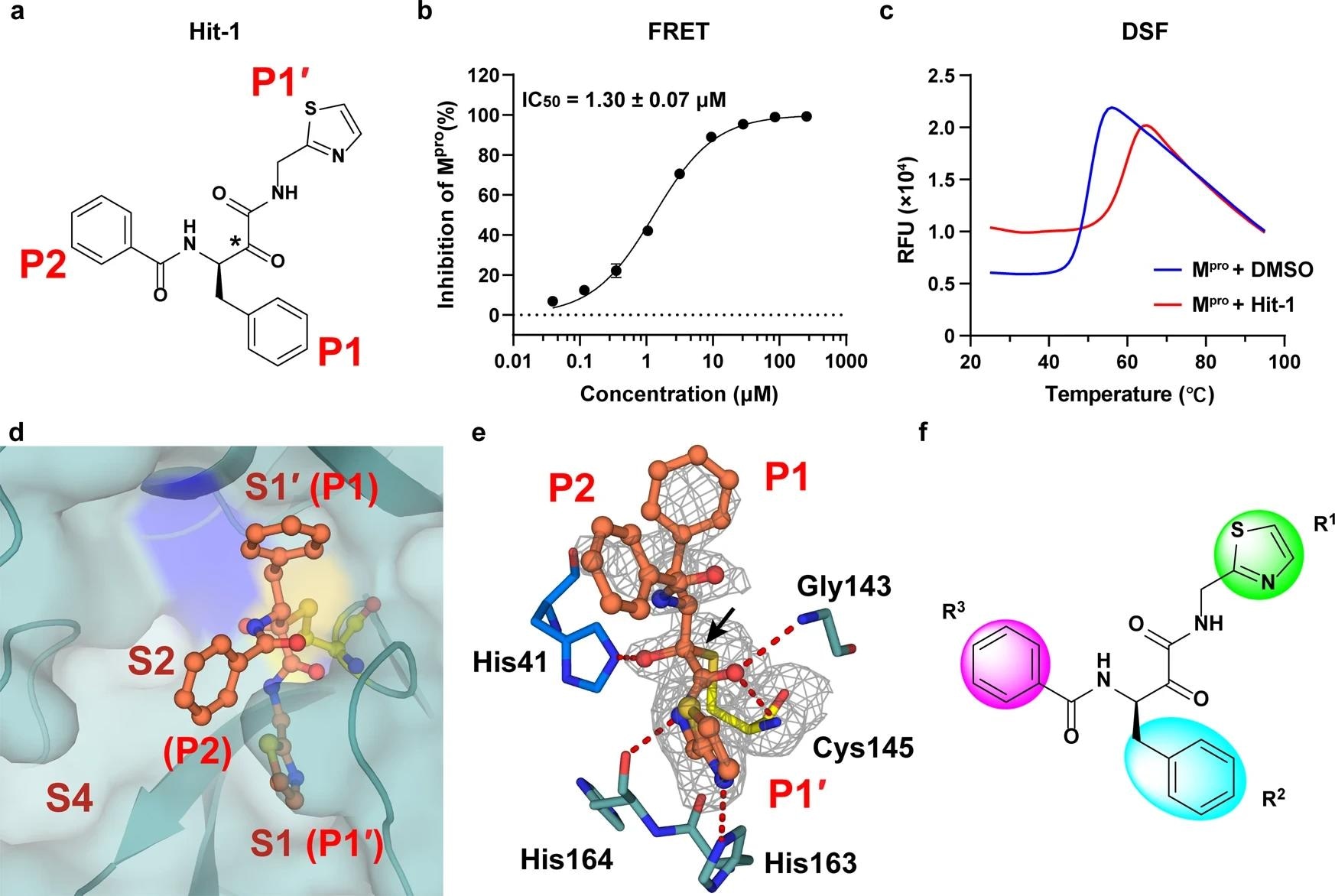 Discovery of a hit compound against SARS-CoV-2 Mpro. a The chemical structure of Hit-1. The P1′, P1, and P2 moieties of Hit-1 are labeled. The warhead carbon is marked with a black asterisk. b Dose-activity curve of Hit-1 against SARS-CoV-2 Mpro in the FRET assay. Data shown are the mean ± standard deviation (SD) from three independent experiments. c Differential scanning fluorimetry analysis of the effect of Hit-1 on SARS-CoV-2 Mpro stability. Exposure of hydrophobic residues monitored by an increase in relative fluorescence units (RFUs). Curves represent the average of three experiments. d Hit-1 (orange) is located at the substrate-binding pocket of Mpro (cyan). His41 of Mpro is in blue, Cys145 is yellow. Pockets (S1′, S1, S2, and S4) of Mpro and moieties (P1′, P1 and P2) of Hit-1 are both labeled. e Interactions between Hit-1 and Mpro. Fo – Fc density map is shown for Hit-1 (gray mesh, σ = 2.5) and Cys145. Covalent bond is shown by a black arrow and hydrogen bonds are displayed by red dashed lines. f Regions of Hit-1 for structural optimization. Images in d, e were processed by using PyMOL (https://pymol.org)
Discovery of a hit compound against SARS-CoV-2 Mpro. a The chemical structure of Hit-1. The P1′, P1, and P2 moieties of Hit-1 are labeled. The warhead carbon is marked with a black asterisk. b Dose-activity curve of Hit-1 against SARS-CoV-2 Mpro in the FRET assay. Data shown are the mean ± standard deviation (SD) from three independent experiments. c Differential scanning fluorimetry analysis of the effect of Hit-1 on SARS-CoV-2 Mpro stability. Exposure of hydrophobic residues monitored by an increase in relative fluorescence units (RFUs). Curves represent the average of three experiments. d Hit-1 (orange) is located at the substrate-binding pocket of Mpro (cyan). His41 of Mpro is in blue, Cys145 is yellow. Pockets (S1′, S1, S2, and S4) of Mpro and moieties (P1′, P1 and P2) of Hit-1 are both labeled. e Interactions between Hit-1 and Mpro. Fo – Fc density map is shown for Hit-1 (gray mesh, σ = 2.5) and Cys145. Covalent bond is shown by a black arrow and hydrogen bonds are displayed by red dashed lines. f Regions of Hit-1 for structural optimization. Images in d, e were processed by using PyMOL (https://pymol.org)
The potency of Hit-1 was improved via stepwise structural optimization, particularly at three regions, namely, thiazole, benzyl, and phenyl. As a result, several candidates were synthesized, which were analyzed based on PK properties, safety, oral bioavailability, cytotoxicity, and stability to obtain the best antiviral agent against SARS-CoV-2 variants. Among all the synthesized candidates, SY110 was the most potent Mpro inhibitor, with an IC50 of 14.4 nM against Mpro.
The oral treatment of SY110 in K18-hACE2 mice with Omicron infection revealed improved pathological damage in both lungs and turbinate. In addition, SY110 demonstrated outstanding in vitro safety profiles in the Ames test, chromosome aberration test, Cytochrome P450 (CYP) assay, and human ether-à-go-go-related gene (hERG) test. In rates, the maximum tolerated dose of SY110 was found to be 1.0 g/kg, with no adverse effect.
SY110 exhibited outstanding PK properties in rats, dogs, mice, and cynomolgus monkeys, with good oral bioavailability. In addition, this compound showed significant antiviral effects against the Alpha, Beta, and Omicron BA.2 and BA.5 sublineages. Importantly, SY110 overcame the Nirmatrelvir-resistances caused by mutations at E166N and E166V.
Conclusions
The current study highlighted the potential of SY110 as a pan-coronavirus antiviral inhibitor, including SARS-CoV-2. When administered orally, this Mpro inhibitor revealed favorable PK properties, excellent oral bioavailability, and a considerable safety profile. In the future, the clinical therapeutic efficacy of SY110 needs to be further validated through clinical trial studies. The authors expressed the possibility of further improving HLM stability for SY110.