Scientists at the University of Colorado Boulder, USA, have deciphered molecular mechanisms involved in the pathogenesis of Amyotrophic Lateral Sclerosis (ALS). They have identified paternally expressed gene 10 (PEG10) as a significant contributor to disease pathogenesis.
amyotrophic Lateral Sclerosis (ALS) is a life-threatening neurodegenerative disease characterized by progressive loss of motor neuron functions, leading to impaired voluntary muscle movement. Sporadic and familial ALS accounts for 90% and 10% of all ALS cases, respectively.
In familial ALS, mutations in a variety of genes, including the proteasome shuttle factor Ubiquilin-2 (UBQLN2), have been identified. In animals with UBQLN2-induced familial ALS, a high abundance of domesticated retrotransposon PEG10 has been observed. Domesticated retrotransposons are a class of genes that encode virus-like proteins that cannot replicate and have evolved adaptive functions.
PEG10 is a domesticated gag-pol retrotransposon that plays crucial roles in placental development and cancer progression. This gene codes for both gag and pol domains separated by a programmed ribosome frameshifting site.
In the current study, scientists have investigated the involvement of PEG10 in the pathogenesis of ALS.
Role of UBQLN2
UBQLN2 is a proteasome shuttle factor highly expressed in neural and muscle tissues. In familial ALS, a mutation in the small, proline-rich PXX repeat region of UBQLN2 is frequently observed.
To investigate the association between UBQLN2 and PEG10, scientists used human embryonic stem cells lacking UBQLN1, UBQLN2, or UBQLN4 genes and measured the level of PEG10. The findings revealed that only UBQLN2-lacking stem cells have increased accumulation of gag-pol form of PEG10, indicating that PEG10 is an exclusive substrate of UBQLN2.
In cells expressing UBQLN2 mutants, significantly higher levels of gag-pol PEG10 were detected compared to that in cells expressing wild-type UBQLN2. This indicates that mutations in UBQLN2 are associated with the loss of its proteasomal degradation function. Further experimentation revealed that the C-terminal polyproline region of PEG10 is required for its degradation by UBQLN2.
Self-processing of PEG10 gag-pol protein
PEG10 is known to undergo the self-cleavage process to generate protein fragments with distinct functions. In this study, scientists specifically showed that the self-cleavage of PEG10 at the N-terminal region leads to the splitting of the capsid region. Similarly, the C-terminal cleavage resulted in the generation of a zinc finger containing fragments reminiscent of retrotransposon and retroviral nucleocapsids. These cleavages were mediated by the retroviral aspartic protease domain of PEG10.
By tracking self-cleavage-generated protein fragments through confocal microscopy, scientists observed that only nucleocapsid fragment is uniquely localized to the nucleus. RNA sequencing and cluster profiling of differentially expressed genes revealed that upon nuclear translocation, the nucleocapsid fragment induces transcriptional changes similar to that caused by the full-length gag-pol protein.
By conducting pathway analysis of differentially expressed genes, scientists observed that the nucleocapsid fragment strongly changes the expression of genes involved in axon guidance and extension. Notably, they observed that the nucleocapsid fragment significantly increases the expression of an axon remodeling gene DCLK1.
By analyzing gene expression profiles of a large cohort of post-mortem ALS patient spinal cord samples, they observed similarly increased levels of DCLK1 transcripts, highlighting the involvement of this transcriptional alteration in ALS pathogenesis.
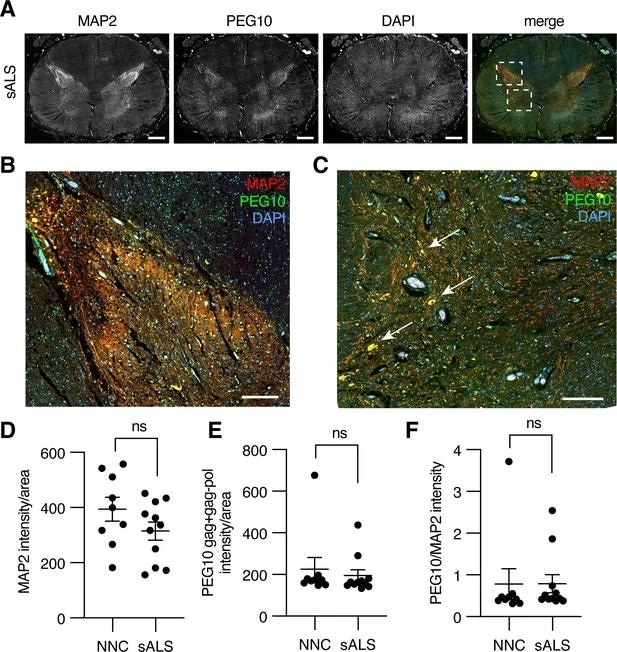
Paternally expressed gene 10 (PEG10) is enriched in horns of the lumbosacral spinal cord. (A) Tiled image of full-thickness lumbosacral spinal cord section demonstrating Microtubule-associated protein 2 (MAP2), PEG10, and DAPI staining. Merged image shows outlines of boxes for closer examination in (B) and (C). Scale bar 1 mm. (B) Merged image of the posterior horn of the spinal cord showing enrichment for both MAP2 and PEG10 signals. Scale bar 200 μm. (C) Merged image of the anterior horn of the spinal cord showing colocalization of MAP2 and PEG10 signal in cell bodies (arrows). Scale bar 200 μm. (D–E) Quantitation of (D) MAP2 and (E) PEG10 signal in spinal cord sections. Signal intensity of MAP2 and PEG10 were determined per mm2 of spinal cord section for a cohort of non-neurological controls (NNC, n=9) and sporadic Amyotrophic Lateral Sclerosis (ALS) patients (sALS, n=11). (F) Relative PEG10 per MAP2 intensity was quantified for each section, to account for potential loss in neuronal staining due to ALS. No significant differences in intensity or localization were observed in the disease condition. For (D–F) mean ± SEM is shown.
PEG10 abundance in human ALS tissues
Scientists conducted a global proteomic analysis using a tandem mass tagging approach for liquid-chromatography mass spectrometry to assess the abundance of PEG10 in post-mortem spinal cord tissue samples isolated from ALS patients and control subjects without any known neurological disease.
They observed significantly elevated levels of PEG10 in ALS samples compared to that in control samples. The abundance of PEG10 in ALS samples was associated with a reduction of neurogranin and neurofilament, known biomarkers of neurological disease.
Study significance
The study reveals that genetic mutations in UBQLN2 are responsible for PEG10 abundance in ALS tissues. PEG10 undergoes protease-dependent self-cleavage to generate a nucleocapsid fragment, which translocates to the nuclease and alters the expression of genes involved in axon remodeling.
Overall, the study establishes a link between PEG10 dysregulation and the development of ALS.
Journal reference:
- Holly H Black, Jessica L Hanson, Julia E Roberts, Shannon N Leslie, Will Campodonico, Christopher C Ebmeier, G Aaron Holling, Jian Wei Tay, Autumn M Matthews, Elizabeth Ung, Cristina I Lau, Alexandra M Whiteley (2023) UBQLN2 restrains the domesticated retrotransposon PEG10 to maintain neuronal health in ALS eLife 12:e79452, https://doi.org/10.7554/eLife.79452, https://elifesciences.org/articles/79452