Studies have identified ~3,000 differentially methylated positions (DMPs) in psoriasis and PsA patients. Typically, chronic joint inflammation triggering PsA manifests within 10 years after the onset of skin psoriasis; however, in nearly 15% of patients, PsA is the initial presentation, which leads to diagnostic delays.
Even though the complex pathophysiology of psoriasis/PsA remains incompletely understood, studies have established the role of epigenetic dysregulation involving effector CD4+ and CD8+ T-cells and the tumor necrosis factor (TNF)/interleukin (IL-)23/IL-17 cytokine axis in their pathophysiology.
It is a key pathway in all T-cell-mediated chronic autoimmune/inflammatory diseases, including psoriasis and PsA. T helper 17 (Th17) cells are a unique Th-cell subset known to secrete IL-17 and other key inflammatory effector cytokines involved in psoriasis. IL-23 promotes the survival and maintenance of Th17 cells; accordingly, researchers have observed its higher expression in psoriatic skin lesions.
Studies have also shown that the blockade of IL-17/23 is an effective treatment for psoriasis/PsA. Likewise, a recently identified Th22 effector Th-cell cell subset, characterized by IL-22/13 production, has been implicated in psoriasis.
Overall, therapeutic interventions targeted at correcting altered DNA methylation patterns in psoriasis/PsA might help permanently resolve immune dysregulation associated with psoriasis/PsA.
About the study
In the present study, researchers collected peripheral blood mononuclear cells (PBMCs) from 12, eight, and eight patients with chronic plaque psoriasis, PsA, and healthy controls (HCs), respectively, separated their CD4+ T-cells through fluorescence-activated cell sorting (FACS) sorting and performed DNA methylation profiling.
Further, they performed Bioinformatic Gene Ontology (GO) enrichment and KEGG pathway analyses to identify precise biological processes associated with altered DNA methylation patterns. They consulted the Interferome database to calculate DNA Methylation Scores, indicating genes under the control of interferon (IFN).
Further, the team performed supervised Partial Least-Squares Discriminant Analysis (PLS-DA) to identify methylation signatures that distinguished "all" psoriasis patients and HCs. They also performed differentially methylated regions (DMRs) analysis in the sub-cohorts of the study.
DMRs are tissue-specific components of contiguous differentially methylated Cytosine and Guanine separated by phosphate (CpG) sites. Studies have implicated DMRs in disease stages and outcomes of several autoimmune disorders, such as rheumatoid arthritis (RA), Sjögren's syndrome, and systemic lupus erythematosus (SLE).
Since all study participants received cytokine-blocking agents, e.g., IL-17 or TNF inhibitors, the team focused on DNA methylation analyses of DMPs in genes involved in the IL-17/TNF pathway. This approach used Psoriasis Area and Severity Index (PASI) scores to show the correlation between DNA methylation and skin disease activity.
Results
The numbers and proportions of CD4+ T-cells and their subsets did not differ across psoriasis, PsA, and HCs, suggesting the observed differences among cohorts were due to varying DNA methylation profiles of CD4+ T-cells.
In addition, the authors noted altered DNA methylation affecting cell junction assembly gene in CD8+ T-cells of "all" psoriasis patients compared to HCs, suggesting that these alterations likely affected biophysical skin properties, e.g., neutrophils recruitment to the skin epidermis.
Samples from skin psoriasis versus PsA patients clustered independently in supervised PLS-DA analysis, and 20 DMPs in PLS-DA component 1 covering three CpG sites were most predictive of psoriasis.
First was the cg01877366 site in the Trafficking Protein Particle Complex Subunit 9 (TRAPPC9), a gene involved in the nuclear factor (NF)-κB activation. A previous study identified TRAPPC9 as a candidate gene in PsA patients not responding to TNF inhibitors. Second was the cg1120622 site in the reversionless 3-like (REV3L) gene, identified as a candidate for gene therapy for psoriasis and PsA.
The third CpG site was the cg18925478 in Phosphatase and Actin Regulator 2 (PHACTR2) gene encoding a protein previously linked to inflammatory bowel disease (IBD). Another study suggested that the PHACTR2 protein was a potential biomarker of SLE exacerbation.
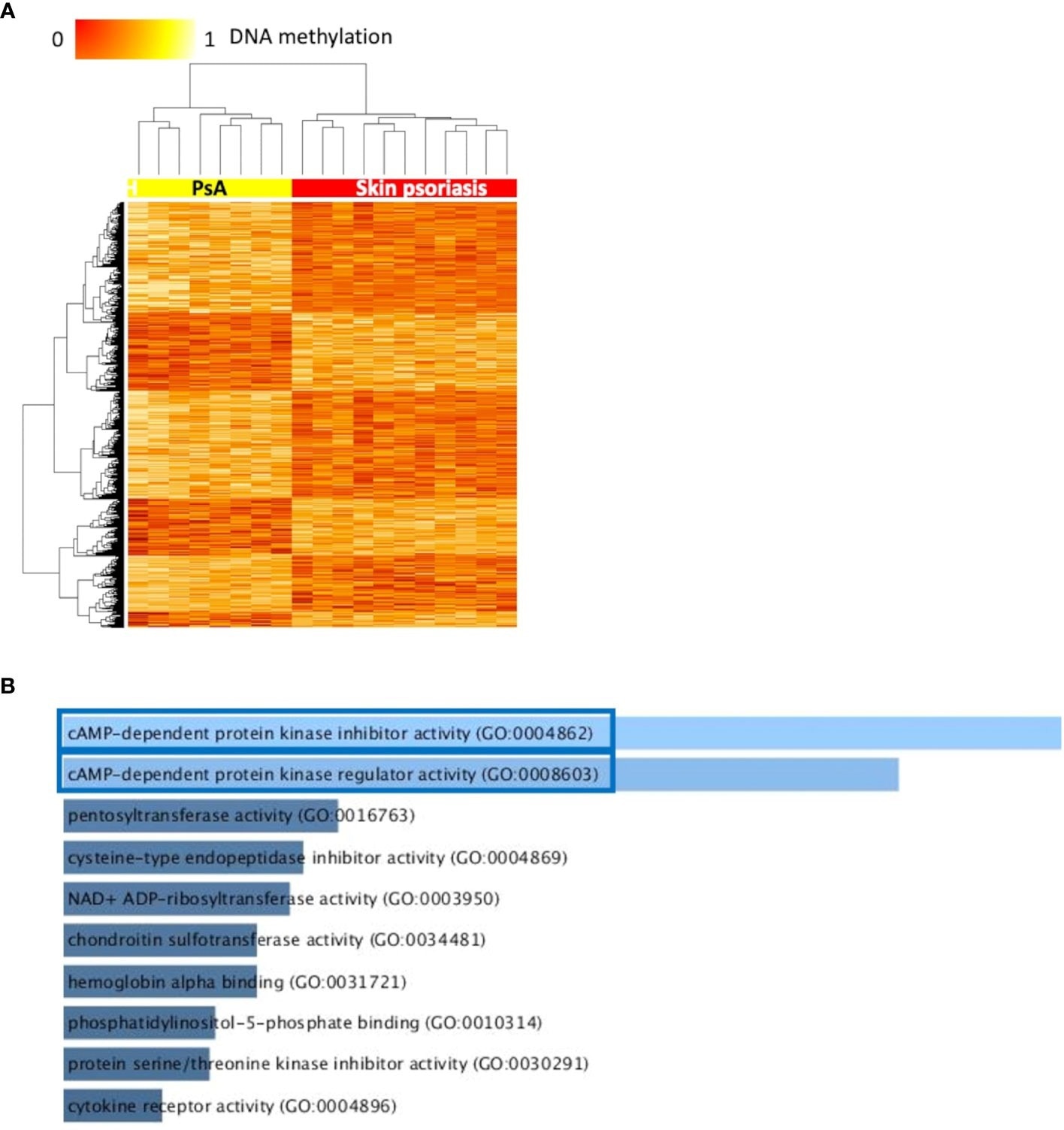 Differentially methylated CpGs differentiate skin psoriasis from PsA. (A) Heat map displaying differentially methylated positions (DMPs) between skin psoriasis and PsA (FDR < 0.05, |Δβ| > 0.1). Normalized DNA methylation levels are displayed on the top left with red indicating reduced methylation and yellow indicating increased methylation levels. (B) Bar diagrams depict the results of Gene Ontology (GO) analysis of hypomethylated genes which presented at least one DMP in their promoter. Top 10 GO terms are represented, and the statistically significant pathways are framed in blue.
Differentially methylated CpGs differentiate skin psoriasis from PsA. (A) Heat map displaying differentially methylated positions (DMPs) between skin psoriasis and PsA (FDR < 0.05, |Δβ| > 0.1). Normalized DNA methylation levels are displayed on the top left with red indicating reduced methylation and yellow indicating increased methylation levels. (B) Bar diagrams depict the results of Gene Ontology (GO) analysis of hypomethylated genes which presented at least one DMP in their promoter. Top 10 GO terms are represented, and the statistically significant pathways are framed in blue.
Consistent with previous reports, nearly 2/3s of DMP-associated genes in this study were involved in the dysregulation of type I/II IFNs or both; thus, DNA methylation scores considering IFN-associated genes allowed disease progression monitoring.
Furthermore, results showed significant enrichment of GO terms related to cyclic adenosine monophosphate (cAMP)-dependent protein kinase (PKA) activity.
The cAMP-PKA pathways regulate the activity and expression of the cAMP-response element modulator (CREM) protein family in CD4+ T-cells, linked to effector cytokine expression in psoriasis/PsA and other autoimmune diseases, such as SLE.
Of all DMRs identified in this study, growth differentiation factor 7 (GDF7) on chromosome 2 and phosphatidylinositol glycan anchor biosynthesis class Z (PIGZ)/piwi-interacting RNA (piRNA) on chromosome 3 were shared in all the comparisons of HCs vs. psoriasis vs. PsA patients.
Studies have suggested that reduced function of the GDF7 gene might be linked to impaired T regulatory (Treg) cell function. Metabolites of glycosylphosphatidylinositol (GPI)-anchor biosynthesis is altered in psoriasis patients, and the PIGZ gene encodes a mannosyl-transferase involved in GPI-anchor biosynthesis.
Another DMP separating psoriasis/PsA patients from HC was dual specificity phosphatase 22 (DUSP22), encoding the c-Jun NH2-terminal kinase (JNK) signaling pathway activator protein that gets dysregulated in psoriasis.
In this study, the authors identified 1,996 DMPs in treatment- naïve and PsA patients receiving biologic disease-modifying antirheumatic drugs (DMARDs), indicating a significant impact of these agents on the DNA methylation landscape.
Remarkably, this was associated with the enrichment of glutathione (GSH) metabolism genes, the most abundant endogenous antioxidant crucial for effector T-cell functions. A recent randomized clinical trial showed that GSH metabolism might be targeted directly by future therapeutic interventions for T-cell-mediated autoimmune diseases, including psoriasis.
Conclusions
The current study revealed distinct DNA methylation profiles in CD4+ T-cells of psoriasis and PsA patients and HCs, highlighting the importance of epigenetic mechanisms in the complex pathophysiology of psoriasis/PsA.
Even though these DNA methylation signatures showed promise as robust and reliable diagnostic/prognostic biomarkers and future treatment targets for psoriasis/PsA, challenges remain in predicting disease progression from psoriasis to PsA and diagnosing PsA in cases with no skin manifestations.
Thus, their application in "real-world" clinical practice needs prospective validation in large-scale independent cohorts.
Journal reference:
- Source: DNA methylation patterns in CD4+ T-cells separate psoriasis patients from healthy controls, and skin psoriasis from psoriatic arthritis, Valentina Natoli, Amandine Charras, Sigrun R. Hofmann, Sarah Northey, Susanne Russ, Felix Schulze, Liza McCann, Susanne Abraham, Christian M. Hedrich, Front. Immunol., 15 August 2023 Sec. Inflammation, Volume 14 - 2023 | doi: https://doi.org/10.3389/fimmu.2023.1245876, https://www.frontiersin.org/articles/10.3389/fimmu.2023.1245876/full