In a recent study published in the journal Nature Biomedical Engineering, a group of researchers demonstrated the use of deep learning to resurrect antibiotic peptides from extinct organisms, providing new solutions for antibiotic resistance and other biomedical challenges.
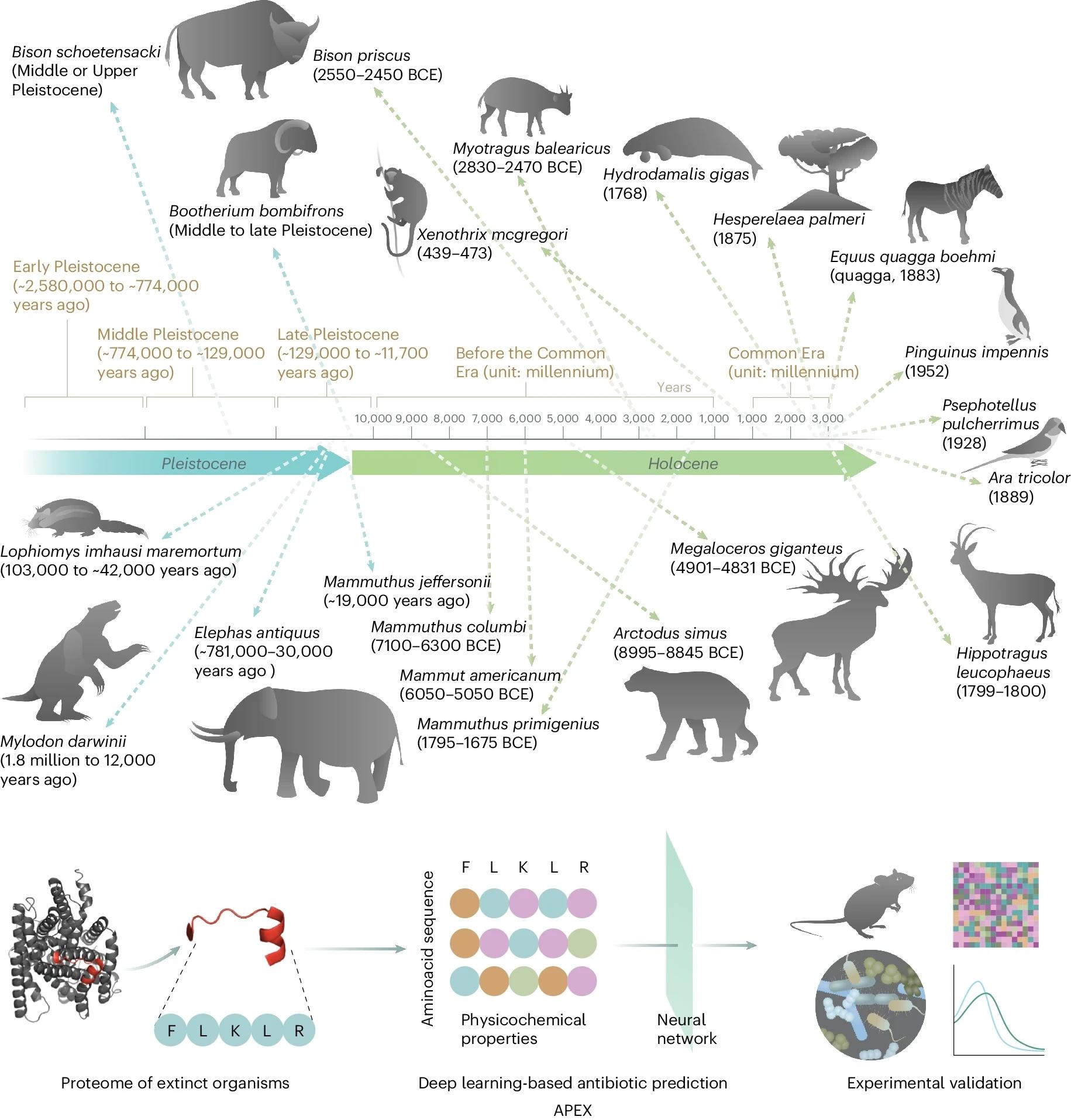 Molecular de-extinction of antibiotics from ancient proteomes using deep learning. All available proteomes of extinct organisms were mined by APEX, our deep learning algorithm. Amino acid sequences ranging from 8 to 50 amino acid residues within proteins from extinct organisms were inputted into multitask deep learning models that trained on both public and in-house peptide data to evaluate the potential antimicrobial activity. The highest-ranked peptides based on predicted antimicrobial activities were then selected and thoroughly characterized against clinically relevant pathogens both in vitro and in animal models. The mechanism of action, physicochemical features, and synergistic interactions of these peptides were also assayed. The dates report the approximate extinction date or period for the organisms studied. The protein and peptide structures shown in the figure were created with PyMOL Molecular Graphics System, version 2.1 Schrödinger, LLC. Study: Deep-learning-enabled antibiotic discovery through molecular de-extinction.
Molecular de-extinction of antibiotics from ancient proteomes using deep learning. All available proteomes of extinct organisms were mined by APEX, our deep learning algorithm. Amino acid sequences ranging from 8 to 50 amino acid residues within proteins from extinct organisms were inputted into multitask deep learning models that trained on both public and in-house peptide data to evaluate the potential antimicrobial activity. The highest-ranked peptides based on predicted antimicrobial activities were then selected and thoroughly characterized against clinically relevant pathogens both in vitro and in animal models. The mechanism of action, physicochemical features, and synergistic interactions of these peptides were also assayed. The dates report the approximate extinction date or period for the organisms studied. The protein and peptide structures shown in the figure were created with PyMOL Molecular Graphics System, version 2.1 Schrödinger, LLC. Study: Deep-learning-enabled antibiotic discovery through molecular de-extinction.
Background
With antimicrobial-resistant infections causing approximately 1.27 million deaths annually worldwide and projections indicating a potential 10 million annual fatalities by 2050, urgent measures are required to combat antibiotic resistance. Additionally, by 2030, around 24 million individuals could face extreme poverty due to the high cost of treating these infections. Molecular de-extinction involves resurrecting extinct molecules to address contemporary challenges such as antibiotic resistance. This approach uncovers new sequence spaces, expanding our understanding of molecular diversity and potential therapeutic designs. Recent computational and artificial intelligence methods have accelerated antibiotic discovery, including through proteome mining. Further research is needed to fully explore the therapeutic potential and safety of resurrected antibiotic peptides and to develop them into effective treatments for combating antibiotic-resistant infections.
About the study
In the present study, researchers collected proteomes of extinct organisms from the National Center for Biotechnology Information (NCBI) taxonomy browser, retrieving 12,860 protein sequences from 208 extinct species. For the modern human proteome, they obtained 20,388 reviewed Homo sapiens proteins from UniProt. An in-house peptide dataset with 14,738 antimicrobial activity measurements from 988 peptides against 34 bacterial strains was used to train and evaluate the Antibiotic Peptide de-Extinction (APEX) model, augmented by publicly available Antimicrobial Peptide (AMP) sequences from the Database of Antimicrobial Activity and Structure of Peptides (DBAASP).
The APEX model architecture featured a recurrent neural network (RNN) with attention layers to process peptide sequences and extract features. Fully connected neural networks (FCNNs) predicted antimicrobial activity and classified AMP/non-AMP labels. Training used mini-batch optimization with the Adam optimizer. Hyperparameters were tuned via grid search and fivefold cross-validation (CV), with ensemble learning averaging predictions from the top eight APEX models.
Researchers screened Encrypted Peptide (EP) sequences from extinct organisms for antimicrobial activity, selectivity, and diversity, resulting in 3,784 unique candidate EPs. They synthesized and validated 69 peptides. Antibacterial assays, membrane permeability, and cytoplasmic membrane depolarization assays assessed peptide activity, while cytotoxicity was evaluated using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays. Resistance to proteolytic degradation was tested in human serum, and secondary structures were analyzed via circular dichroism.
Mouse models for skin abscess and thigh infections tested in vivo efficacy. Statistical analyses were conducted using one-way Analysis of Variance (ANOVA) and GraphPad Prism and Python for data analysis.
Study results
The training dataset included 988 in-house peptides and 5,093 antimicrobial and 5,500 non-antimicrobial peptides from DBAASP. The in-house dataset contained 14,738 antimicrobial activity values from 34 bacterial strains. The dataset was split into a CV set and an independent set. Hyperparameters were tuned using fivefold CV, and the independent set evaluated the final prediction performance.
APEX outperformed baseline machine learning models (elastic net, extra-trees regressor, linear support vector regression, random forest, and gradient boosting decision tree) on most pathogen-specific Minimum Inhibitory Concentration
(MIC) predictions. An ensemble learning approach, averaging predictions from the top eight APEX models, further improved performance.
To compare the predictive power of APEX to a scoring function, researchers tested 49 peptides predicted by the scoring function and 69 peptides predicted by APEX from extinct organisms. Among the 69 APEX-predicted peptides, 21 were derived from secreted proteins and 48 from non-secreted proteins. The peptides were synthesized and experimentally tested for antimicrobial activity against 11 bacterial pathogens, revealing a hit rate of 59% for identifying peptides with antimicrobial activity, significantly higher than the 24% hit rate of the scoring function.
The researchers also assessed the secondary structures of these peptides, finding that APEX-identified peptides predominantly formed α-helical structures, enhancing their membrane interaction and antimicrobial effectiveness. The mechanism of action was investigated through membrane depolarization and permeabilization assays, showing that APEX-predicted peptides effectively depolarized bacterial membranes.
In vivo tests in mouse models of skin abscess and thigh infection showed that several peptides, including AEPs (archaeic EPs) and MEPs (modern EPs), exhibited significant anti-infective efficacy. Notably, peptides such as elephasin-2, hydrodamin-1, megalocerin-1, mammuthusin-2, and mylodonin-2 reduced bacterial loads by 2-5 orders of magnitude, comparable to the widely used antibiotic polymyxin B.
Conclusions
To summarize, this study highlights the successful application of deep learning in predicting antimicrobial activity from peptide sequences, specifically through the development and use of the APEX model. APEX, trained on both in-house and publicly available datasets, utilizes a multitask learning architecture to predict the antimicrobial properties of peptides. The model outperformed traditional machine learning methods in predicting species-specific antimicrobial activity, demonstrating substantial predictive power. The findings underscore the potential of molecular de-extinction for discovering novel therapeutic molecules and addressing antibiotic resistance.