The diagnosis of cancers has traditionally depended on the examination of histopathological preparations of hematoxylin and eosin slides using light microscopes. The advances in digital technology and computational pathology have replaced this with whole-slide images that can be examined computationally, making this form of diagnosis a part of routine clinical practice.
The use of artificial intelligence (AI) in the diagnosis and characterization of cancers using digitized whole-slide images has also grown substantially, with initial efforts focused on improving workflows. However, recent studies have explored a subfield where AI is used extensively to analyze whole-slide images to reveal more than just diagnostic information, including therapeutic responses and prognosis. This is also decreasing the reliance on genomic testing and immunohistochemistry-based methods for cancer diagnosis.
About the study
In the present study, the researchers discussed the largest foundational model developed to date, Virchow. They demonstrated its use in predicting cancer biomarkers across a wide range of common and rare cancers.
Foundational models are large-scale neural networks trained on very large datasets using self-supervised learning. These models create data representations known as embeddings, which can gather generalized data from large datasets to be applied in cases with inadequate data and for predictive tasks such as determining clinical outcomes, genomic changes, and therapeutic responses.
An efficient foundational model can capture broad-spectrum patterns such as tissue architecture, nuclear morphology, cellular morphology, necrosis, staining patterns, neovascularization, inflammation, and expression of biomarkers that can be used to predict various whole-slide image characteristics.
Here, the researchers discussed Virchow, the largest foundational model developed to date, named after the pioneering modern pathologist Rudolf Virchow. The model has been trained on a sizable dataset of nearly 1.5 million hematoxylin and eosin whole-slide images obtained from a hundred thousand patients registered at the Memorial Sloan Kettering Cancer Center (MSKCC). The dataset consists of benign and malignant tissue samples obtained from resections and biopsies of 17 tissue types.
Virchow is a vision transformer model comprising 632 million parameters. It is trained using a self-supervised algorithm that uses local and global regions of the tissue tiles to create embeddings of whole-slide images that can be used for predictive tasks.
To highlight the clinical applications and utility of such a large foundational model, the researchers used the Virchow embeddings created from the large dataset of whole-slide images to train a pan-cancer model and assess its performance in predicting common and rare forms of cancer at the specimen level across various tissues.
The study compared the performance of the Virchow embeddings against Phikon, UNI, and CTransPath embeddings and evaluated the utility of Virchow embeddings along two categories. The first was the performance of the pan-cancer detection model trained using Virchow embeddings on a test dataset consisting of a mix of datasets from MSKCC and external sources spanning seven rare and nine common types of cancers. The effectiveness of Virchow embeddings in biomarker prediction using data from cancers such as lung, bladder, breast, and colon cancers was also evaluated.
Results
The study showed that Virchow embeddings demonstrated the two-fold value of a foundational model of pathology by being generalizable and providing training data efficiency. The pan-cancer model trained on Virchow embeddings was able to detect not only the common forms of cancer but also the rarer histological subtypes in the test dataset.
Furthermore, the performance of the pan-cancer model was comparable to that of clinical-grade cancer-detection models and, in cases of some rare cancers, even exceeded that of the clinical models despite having been trained on datasets with fewer tissue-specific labels.
The researchers stated that the model's performance level was especially notable considering that the dataset used to train the pan-cancer model did not undergo the subpopulation enrichment and quality control performed for training commercially and clinically used AI models.
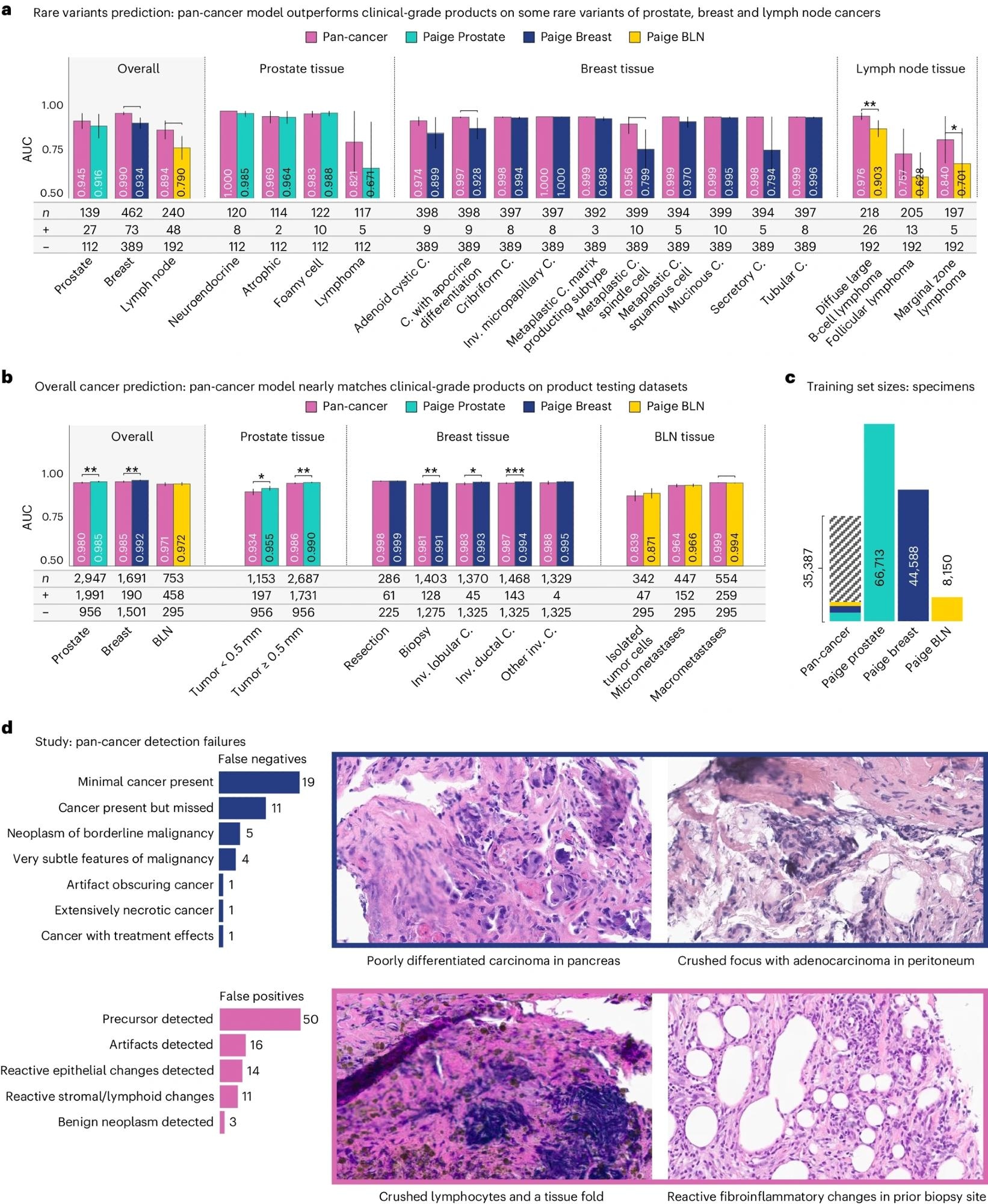 a,b, Performance as measured by AUC of three clinical products compared to the pan-cancer model trained on Virchow embeddings, on the rare variant (a) and product testing datasets (b). The pan-cancer detector, trained on Virchow foundation model embeddings, achieves similar performance to clinical-grade products in general and outperforms them on rare variants of cancers. c, The pan-cancer detector was trained on fewer labeled specimens than the Prostate, Breast and BLN clinical models, including a small fraction of the prostate (teal), breast (blue) and BLN (yellow) tissue specimens that these clinical models were respectively trained on. d, A categorization of failure models of the pan-cancer model and four canonical examples of the primary types of failures. In all panels, * is used to indicate pairwise statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; pairwise DeLong’s test). Error bars denote the two-sided 95% confidence interval, estimated with DeLong’s method. C., carcinoma. Inv., invasive.
a,b, Performance as measured by AUC of three clinical products compared to the pan-cancer model trained on Virchow embeddings, on the rare variant (a) and product testing datasets (b). The pan-cancer detector, trained on Virchow foundation model embeddings, achieves similar performance to clinical-grade products in general and outperforms them on rare variants of cancers. c, The pan-cancer detector was trained on fewer labeled specimens than the Prostate, Breast and BLN clinical models, including a small fraction of the prostate (teal), breast (blue) and BLN (yellow) tissue specimens that these clinical models were respectively trained on. d, A categorization of failure models of the pan-cancer model and four canonical examples of the primary types of failures. In all panels, * is used to indicate pairwise statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; pairwise DeLong’s test). Error bars denote the two-sided 95% confidence interval, estimated with DeLong’s method. C., carcinoma. Inv., invasive.
Conclusions
To conclude, the study showed that a pan-cancer model trained using Virchow embeddings was able to perform comparably and often better than clinical-grade models in detecting common and rare forms of cancer despite being trained on datasets with fewer tissue labels.
Overall, the findings highlighted the significance and utility of foundational models such as Virchow in applications involving limited amounts of training data, providing the basis for various clinical models in cancer pathology.
Journal reference:
- Vorontsov, E., Bozkurt, A., Casson, A., Shaikovski, G., Zelechowski, M., Severson, K., Zimmermann, E., Hall, J., Tenenholtz, N., Fusi, N., Yang, E., Mathieu, P., Eck, van, Lee, D., Viret, J., Robert, E., Wang, Y. K., Kunz, J. D., Matthew, L., & Bernhard, J. H. (2024). A foundation model for clinical-grade computational pathology and rare cancers detection. Nature Medicine. DOI:10.1038/s41591024031410, https://www.nature.com/articles/s41591-024-03141-0