 Study: Brain clocks capture diversity and disparities in aging and dementia across geographically diverse populations. Image Credit: Lightspring / Shutterstock
Study: Brain clocks capture diversity and disparities in aging and dementia across geographically diverse populations. Image Credit: Lightspring / Shutterstock
In a recent study published in the journal Nature Medicine, researchers used deep learning to analyze the impact of geographical, sociodemographic, socioeconomic, neurodegeneration-related, and gender diversity on brain-age gaps across 15 countries. They found that structural socioeconomic inequality, pollution, and health disparities are key predictors of increased brain-age gaps, particularly in Latin American and Caribbean (LAC) regions, with larger gaps observed in females and individuals with cognitive impairments like Alzheimer’s disease (AD).
Background
The brain undergoes dynamic changes with age, which are crucial to understanding, especially in relation to disparities and brain disorders like AD. Brain-age models, which measure brain health across various factors, have the potential to capture diversity in aging but have been underexplored in underrepresented populations like those in LAC. These populations face significant socioeconomic and health disparities, which may impact brain aging. Research on brain aging has mainly focused on populations from the Global North and often uses structural magnetic resonance imaging (MRI), neglecting brain network dynamics captured by functional MRI (fMRI) and electroencephalograms (EEG). While EEG is a more accessible tool in resource-limited settings, its use in large-scale studies is limited by challenges in standardization and integration with fMRI. There is a need to develop scalable brain-age markers using deep learning that incorporate these techniques and account for demographic diversity, especially in underrepresented populations. Therefore, researchers in the present study used graph convolutional networks to predict brain-age gaps and examine the impact of diversity, including geographical, sociodemographic, and health factors, on brain aging.
About the study
The study analyzed resting-state fMRI and EEG datasets of 5,306 participants across 15 countries in the LAC and non-LAC regions. fMRI data were collected from 2,953 participants in Argentina, Chile, Colombia, Mexico, Peru, the United States of America, China, and Japan, while EEG data were gathered from 2,353 participants in Argentina, Greece, Brazil, Chile, Colombia, Cuba, Ireland, Italy, Turkey, and the United Kingdom. The participants included 3,509 healthy controls and 1,808 with neurocognitive disorders, namely mild cognitive impairment (MCI), AD, or behavioral variant frontotemporal dementia (bvFTD). The data underwent rigorous preprocessing, including normalization, noise correction, and source space estimation. High-order interactions between brain regions were assessed, with data converted into graphs for analysis via graph convolutional networks (GCNs). An approach involving 80% cross-validation and 20% hold-out testing was used. Data augmentation techniques were employed, and the model’s predictive performance was evaluated using goodness of fit (R²) and root mean square error (r.m.s.e). Gradient-boosting models were used to explore the influence of exposome factors on brain-age gaps. Extensive statistical analyses were conducted to validate the findings, including permutation tests and bootstrapping. Data quality was carefully assessed, and the study adhered to strict ethical guidelines.
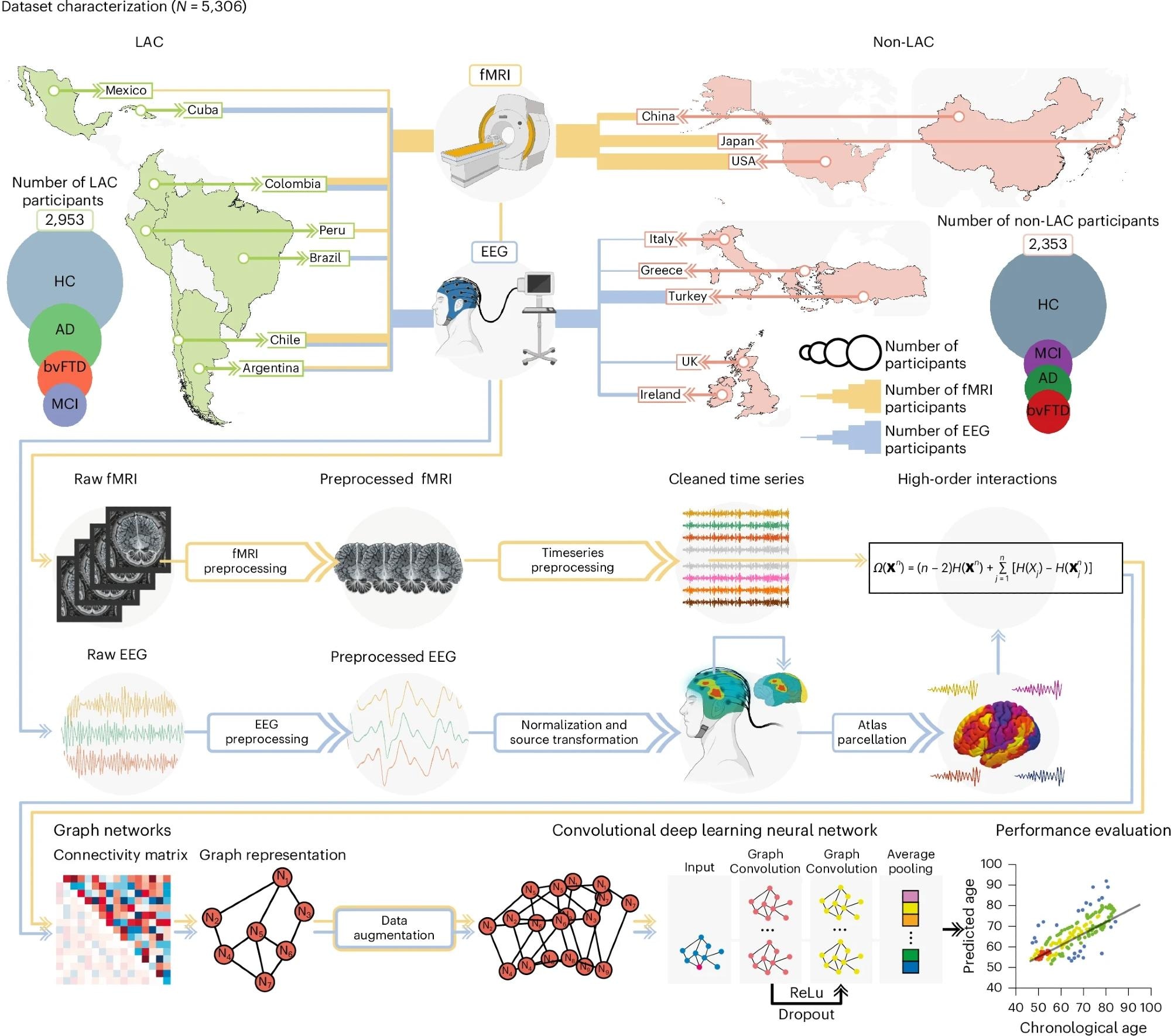 Datasets included LAC and non-LAC healthy controls (HC, total n = 3,509) and participants with Alzheimer disease (AD, total n = 828), bvFTD (total n = 463) and MCI (total n = 517). The fMRI dataset included 2,953 participants from LAC (Argentina, Chile, Colombia, Mexico and Peru) as well as non-LAC (the USA, China and Japan). The EEG dataset involved 2,353 participants from Argentina, Brazil, Chile, Colombia and Cuba (LAC) as well as Greece, Ireland, Italy, Turkey and the UK (non-LAC). The raw fMRI and EEG signals were preprocessed by filtering and artifact removal and the EEG signals were normalized to project them into source space. A parcellation using the automated anatomical labeling (AAL) atlas for both the fMRI and EEG signals was performed to build the nodes from which we calculated the high-order interactions using the Ω-information metric. A connectivity matrix was obtained for both modalities, which was later represented by graphs. Data augmentation was performed only in the testing dataset. The graphs were used as input for a graph convolutional deep learning network (architecture shown in the last row), with separate models for EEG and fMRI. Finally, age prediction was obtained, and the performance was measured by comparing the predicted versus the chronological ages. This figure was partially created with BioRender.com (fMRI and EEG devices).
Datasets included LAC and non-LAC healthy controls (HC, total n = 3,509) and participants with Alzheimer disease (AD, total n = 828), bvFTD (total n = 463) and MCI (total n = 517). The fMRI dataset included 2,953 participants from LAC (Argentina, Chile, Colombia, Mexico and Peru) as well as non-LAC (the USA, China and Japan). The EEG dataset involved 2,353 participants from Argentina, Brazil, Chile, Colombia and Cuba (LAC) as well as Greece, Ireland, Italy, Turkey and the UK (non-LAC). The raw fMRI and EEG signals were preprocessed by filtering and artifact removal and the EEG signals were normalized to project them into source space. A parcellation using the automated anatomical labeling (AAL) atlas for both the fMRI and EEG signals was performed to build the nodes from which we calculated the high-order interactions using the Ω-information metric. A connectivity matrix was obtained for both modalities, which was later represented by graphs. Data augmentation was performed only in the testing dataset. The graphs were used as input for a graph convolutional deep learning network (architecture shown in the last row), with separate models for EEG and fMRI. Finally, age prediction was obtained, and the performance was measured by comparing the predicted versus the chronological ages. This figure was partially created with BioRender.com (fMRI and EEG devices).
Results and discussion
The brain-aging models showed adequate predictive performance. The key predictive brain-regional features were centered around frontoposterior networks, including nodes in the precentral gyrus, middle occipital gyrus, and superior and middle frontal gyri. Additional key nodes for the fMRI model were the inferior frontal gyri, anterior and median cingulate, and paracingulate gyri. For the EEG model, the inferior occipital gyrus and the superior and inferior parietal gyri were also significant.
Notably, when analyzing non-LAC datasets, the models showed similar patterns in predictive features but with a slightly reduced fit. In contrast, models trained on LAC datasets revealed a moderate fit and increased r.m.s.e. values, highlighting biases towards predicting older brain ages, particularly for female participants. Furthermore, the examination of cross-regional effects demonstrated that training with non-LAC data and testing on LAC resulted in positive mean directional errors (MDE), indicating biases towards older brain ages. Additionally, brain-age gaps were observed to be widened in clinical populations, suggesting accelerated aging in conditions like MCI and AD compared to healthy controls.
These findings highlight the complexities of brain aging across different populations. They emphasize the importance of considering diversity factors in neurocognitive assessments. The study is strengthened by using diverse datasets across several countries, integrating fMRI and EEG data, and developing scalable, personalized brain health metrics applicable to diverse and underrepresented populations. However, the study is limited by the lack of clinical EEG data from non-LAC regions, reliance on unimodal brain-age gap measurement, limited regional data, and the absence of individual-level demographic factors like gender identity, socioeconomic status, and ethnicity.
Conclusion
In conclusion, the study demonstrates that brain clock models are sensitive to diverse factors such as geography, sex, macrosocial influences, and diseases, despite data variability. By leveraging deep learning on high-order brain interactions across fMRI and EEG, the research paves the way for inclusive, accessible tools to assess disparities in brain aging. It could potentially aid the identification and staging of neurocognitive disorders like MCI, AD, and bvFTD and support personalized medicine approaches globally.