Alzheimer's disease is characterized by the accumulation and aggregation of amyloid β and tau fibrils that form neurofibrillary tables in the brain. The neurodegeneration and cognitive decline that occur with Alzheimer's disease are caused by the initial formation of amyloid β plaques, which facilitate the formation and spread of neurofibrillary tau tangles into the neocortex.
However, although the roles of amyloid β plaques and tau fibrils are known, Alzheimer's disease is multifactorial and involves numerous other proteins and biological pathways. Recent studies that have conducted proteomic analyses on cerebrospinal fluid have found significant variations in the levels of several proteins across different stages of the disease.
Furthermore, compared to traditional biomarkers such as amyloid β and phosphorylated tau levels, the use of advanced imaging techniques such as PET scans has also provided more accurate insights into the pathology of the brain in Alzheimer's disease.
About the study
In the present study, the researchers combined PET imaging techniques and proteomic analysis of cerebrospinal fluid to measure the aggregation of amyloid β and tau tangles in a group of Alzheimer's patients at different stages of disease pathology. They focused on understanding the proteomic changes that occur in different pathological stages rather than the changes in cognitive symptoms.
The study included 877 participants who had been phenotyped as part of the BioFINDER-2 study, which had conducted a proteomic analysis of cerebrospinal fluid samples from all the participants. The researchers used the amyloid β and tau pathologies to categorize the participants into different groups.
The ratio of amyloid β 42 to amyloid β 40 in the cerebrospinal fluid samples, which is generally used to detect amyloid β plaques in the brain, was used to identify amyloid β pathology. Tau pathology was determined using tau-PET imaging, which can visualize insoluble tau fibrils in the cortex.
The participants were classified into four groups: those with no amyloid β or tau pathologies, those who were amyloid β positive and tau negative, those who were positive for both amyloid β and tau, and those who had a neurodegenerative disease with no amyloid β or tau pathologies.
To determine which proteins were differentially abundant in the early stages of Alzheimer's disease, the scientists first compared the protein levels between the only positive group for amyloid β and the negative group for both amyloid β and tau.
Subsequently, comparisons between the group that was positive only for amyloid β and the group that was positive for both amyloid β and tau revealed the proteins that were associated with the more advanced stages of Alzheimer's disease that manifested tau pathology.
Factors such as overall protein levels, sex, and age were included as covariates in the study. Furthermore, the findings were validated using a separate cohort from the original study population. The Alzheimer's Disease Neuroimaging Initiative was also used to validate the findings further.
Imaging transcriptomics also examined the relationship between amyloid β and tau accumulation and the early differentially expressed proteins. Comparisons of the proteins differentially expressed in Alzheimer's disease with those from other neurodegenerative diseases not involving amyloid β and tau pathologies also helped determine the specificity of these proteins in Alzheimer's disease.
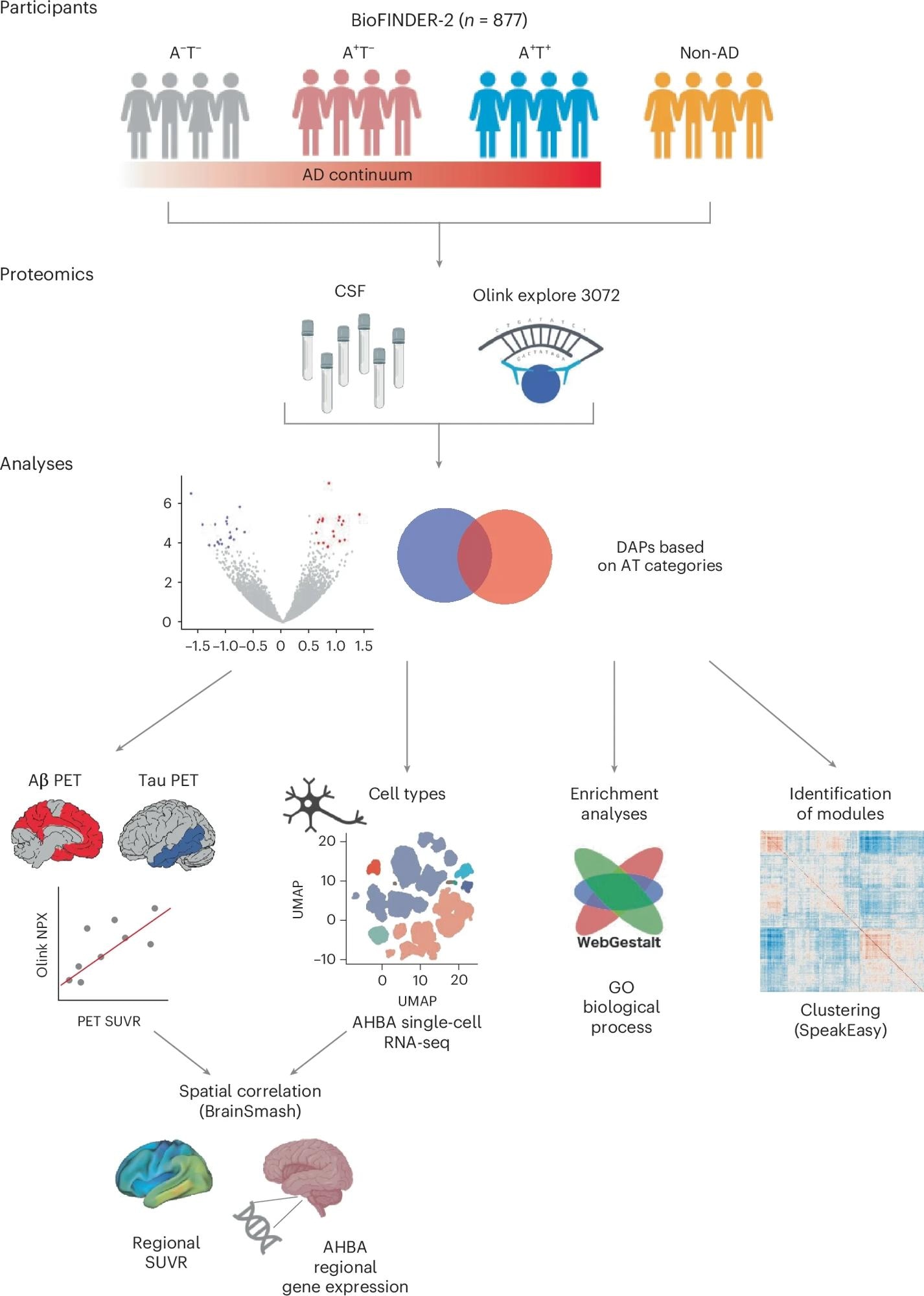 From all BioFINDER-2 participants with Olink proteomic data from CSF, we first assessed DAPs across the different A/T categories. From these DAPs, we then evaluated whether: (1) they were independently related to Aβ plaques or tau tangle pathology load (baseline PET) and rate of change (longitudinal PET); (2) the proteins’ regional gene expression in the brain matched the regional PET pattern; and (3) they were enriched in different cell types or biological processes using enrichment analyses. Last, we derived protein co-expression modules to investigate the overlap between such modules and the DAPs.
From all BioFINDER-2 participants with Olink proteomic data from CSF, we first assessed DAPs across the different A/T categories. From these DAPs, we then evaluated whether: (1) they were independently related to Aβ plaques or tau tangle pathology load (baseline PET) and rate of change (longitudinal PET); (2) the proteins’ regional gene expression in the brain matched the regional PET pattern; and (3) they were enriched in different cell types or biological processes using enrichment analyses. Last, we derived protein co-expression modules to investigate the overlap between such modules and the DAPs.
Results
The study identified 127 differentially abundant proteins across the spectrum of Alzheimer's disease pathology. The proteins related to the amyloid β pathology were expressed mainly in glial cells. The two predominant differentially abundant proteins were SMOC1, a modular calcium-binding protein 1, and integrin subunit alpha M (ITGAM).
Numerous proteins involved in the adenosine triphosphate (ATP) metabolism showed independent associations with tau accumulation and tangles and were differentially abundant in the neurons. Furthermore, the proteome of Alzheimer's disease was identified as distinct since the differentially abundant proteins showed only a 20% overlap with those from other neurodegenerative diseases.
These differentially abundant proteins also showed links to various biological processes. The proteins that were differentially abundant in the early stages of Alzheimer's disease were linked to cellular detoxification and transmission across synapses, while the proteins that were differentially abundant in the later stages of the disease were involved in cellular structure and metabolic processes.
Proteins such as fatty acid-binding protein 3 and enolase 2 were found to be associated only with tau pathology and not with amyloid β pathology, indicating the potential to use these proteins as markers for detecting tau-associated neurodegeneration in Alzheimer's disease.
Conclusions
Overall, the findings comprehensively assessed the differential abundance of proteins in different stages of Alzheimer's disease and highlighted the distinct molecular pathways involved in different disease stages. These findings highlight some of the protein signatures of the different pathologies of Alzheimer's disease and provide potential targets for novel therapeutic strategies.
Journal reference:
- Binette, P., Gaiteri, C., Wennström, M., Kumar, A., Hristovska, I., Spotorno, N., Salvadó, G., Strandberg, O., Mathys, H., Tsai, L., Palmqvist, S., MattssonCarlgren, N., Janelidze, S., Stomrud, E., Vogel, J. W., & Hansson, O. (2024). Proteomic changes in Alzheimer disease associated with progressive Aβ plaque and tau tangle pathologies. Nature Neuroscience. DOI:10.1038/s4159302401737w, https://www.nature.com/articles/s41593-024-01737-w