In order to sustain themselves, living organisms rely on proteins called enzymes to catalyze the necessary organic reactions. This reaction process begins when the substrate (S) reversibly binds to the active site of the enzyme (E) to form an enzyme-substrate complex (E-S) with rate constants of k1 and k-1. The subsequent step is the release of the product (P) with a rate constant of k21 by the enzyme. The equation below shows the general reaction scheme of an enzyme catalyzed reaction.

In order to get a better understanding of the behavior of enzymes, a kinetic description of their activity is vital. The Michaelis-Menten model is one of the best-known models of enzyme kinetics. It is defined by an equation that relates the reaction rate, ν (i.e. the rate of the formation of [P]), to the concentration of the substrate, [S]. The Michaelis-Menten equation is given below:
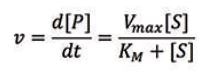
Two important parameters can be determined from the Michaelis-Menten model, Vmax and KM. Vmax represents the maximum rate of product formation at a saturating substrate concentration and is a measure of efficiency of the catalytic properties of the enzyme. The Michaelis constant, KM, represents the concentration of substrate at which the reaction rate is half of Vmax .It is often used to quantify the affinity of the active site for the substrate (the smaller the KM value the higher the affinity).
Typically, Vmax and KM are calculated by first determining the initial reaction rate of an enzyme at a number of different substrate concentrations. A graph of reaction rate against concentration is then plotted, known as Michaelis-Menten plot. By reciprocating both axes on the Michaelis-Menten plot, the Lineweaver-Burk plot can be obtained from which the Vmax and KM can be extracted from the line of best fit.
Procedure
Preparing Stock Solutions
N-acetyl-DL-methionine (0.382 g) was suspended in 2 mL of D2O along with 0.112 g of KH2PO4. Sodium Hydroxide (2 M solution in D2O) was added carefully to bring the pH to 7 using pH paper. The resulting solution is then diluted to 5 mL in a volumetric flask using D2O. The final solution contained 400 mM of N-acetyl-DL-methionine. A stock solution of enzyme is prepared by dissolving 10 mg of porcine acylase and 1.5 mg of CoCl2.6H2O in 10 mL of D2O).
Monitoring the Reaction with the NMReady-60
The solution of N-acetyl-DL-methionine 500 µL is transferred to an NMR tube and 1H NMR spectrum was obtained (spectral width = 20 ppm, spectral center = 5 ppm, number of scans = 16, delay= 0.5 sec, number of points = 4096). The reaction is initiated by adding 100 µL of the enzyme solution to the NMR tube followed by vigorous mixing. A 1H NMR spectrum is recorded every 4 minutes for 2 hours using the kinetics module on the NMReady-60 (wait type = linear, number of clusters = 40, wait units = seconds, wait time (tau) = 160). To monitor the progress of the reaction the integrals of the α-methine protons were measured for the reactant (N-acetyl-DL-methionine, 4.25 ppm and product L-methionine, 3.85 ppm).
Results
Discussion
Figure 1 is the 1H NMR spectrum of the hydrolysis reaction and demonstrates the depletion of the substrate, N-acetyl-DL-methionine (4.25 ppm), as well as the simultaneous appearance of the product L-methione (3.85 ppm). A key point of note is that the signal at 4.25 ppm is never completely erased due to the D-enantiomer of the racemic mixture remaining in the solution without being hydrolyzed by the porcine acylase.
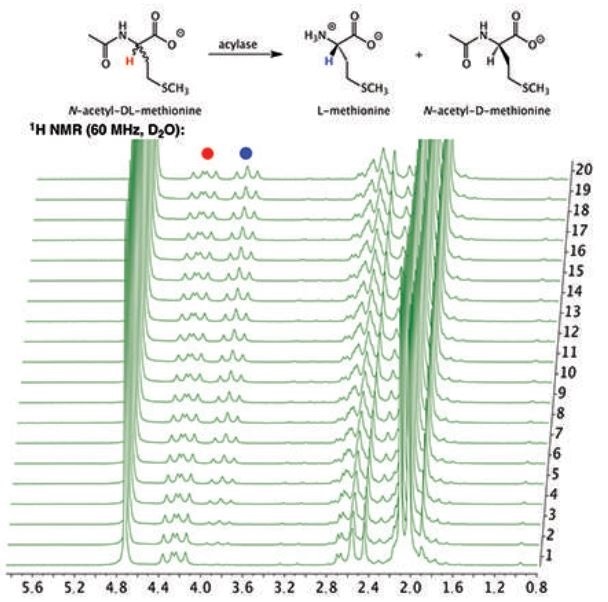
Figure 1. Stacked plot of 1H NMR spectra of the hydrolysis of N-acetyl-DL-methionine by porcine acylase to produce L-methione.
A plot of substrate concentration over time is shown in Figure 2. With the substrate concentration reaching a plateau, it can be seen that the reaction is complete within the hour. The Michaelis-Menten plot in Figure 3 displays the change of reaction rate as a function of substrate concentration. Although the Michaelis-Menten experiment is usually performed by measuring the reaction rate at several initial substrate concentrations, in this case the experiment can be condensed into one reaction.
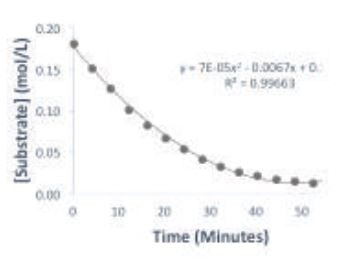
Figure 2. Plot of substrate concentration over time of the reaction.
![Michaelis-Menten plot of the reaction. The data was fitted to: V = (Vmax [S]) / (KM + [S]).](https://www.news-medical.net/image-handler/picture/2019/3/Art1-Pic5.jpg)
Figure 3. Michaelis-Menten plot of the reaction. The data was fitted to: V = (Vmax [S]) / (KM + [S]).
As multiple 1H NMR spectra are acquired as the reaction proceeds, the spectrum allows substrate concentration to be determined and the reaction rate can be approximated by calculating the difference in substrate concentration over a known time interval. Therefore at higher substrate concentration it is seen that the reaction rate begins to reach a plateau which represents the Vmax at this substrate concentration.
From the Michaelis-Menten plot, the Lineweaver-Burk lot (Figure 4) is constructed by reciprocating both axes. Subsequently it was found that the KM = 0.24 mol L-1 and Vmax -0.3152 mmol L-1s-1.
![Lineweaver-Burk plot of the reaction. The data was fitted to the equation 1/V = (KM/Vmax [S]) + 1/Vmax from which the values of KM (0.24 mol L-1) and Vmax (0.3152 mmol L-1 s-1) were extracted.](https://www.news-medical.net/image-handler/picture/2019/3/Art1-Pic6.jpg)
Figure 4. Lineweaver-Burk plot of the reaction. The data was fitted to the equation 1/V = (KM/Vmax [S]) + 1/Vmax from which the values of KM (0.24 mol L-1) and Vmax (0.3152 mmol L-1 s-1) were extracted.
Conclusions
This experiment studied the enzymatic hydrolysis of N-acetyl-L-methionine. Due to the variation in chemical shifts of the α-methine protons in the substrate and product, 1H NMR spectroscopy could be used to determine the progress of the reaction using the Nanalysis 60PRO. Furthermore, quantitative data was obtained from the spectra that allowed a Michaelis-Menten and Lineweaver-Burk plot to be produced which were then used to determine the Vmax and KM values of the enzymatic reaction.
References and Further Reading
- Le H.; Algaze, S.; Tan, E. Michaelis-Menten Kinetics (accessed Dec 4, 2018).
- BIanco, A.; Blanco, G. Medical biochemistry; Academic Press: London, United Kingdom, 2017; pp. 1 53-1 75.
- Berg, J.; Tymoczko, J.; Stryer, L. Biochemistry; 5th ed.; W.H. Freeman and Co.: New York, 2002.
- OIsen, IR., Olsen, J. and Giles, G. "An Enzyme Kinetics Experiment for the Undergraduate Organic Chemistry Laboratory." J. Chem, Educ., 2010, 87(9), pp.956-957.
Data Accessibility
The Nanalysis 60PRO allows data to be processed directly and then printed. The data can also be exported directly to a USB or networked file where it can be worked up using third party NMR processing software.
About Nanalysis

Nanalysis was established in 2009. The company develops and manufactures portable Nuclear Magnetic Resonance (NMR) spectrometers for the laboratory instrumentation market.
In the fall of 2012, the first product was launched: NMReady™, the first fully featured portable NMR spectrometer in a single compact enclosure requiring no liquid helium or any other cryogens.
The NMReady is used by chemical professionals in all types of industries (oil & gas, chemical, pharma, biotech, food processing) as well as government and university labs.
The NMReady enables all chemical trainees to gain first-hand knowledge of NMR as the premier spectroscopic method by using it in a variety of training environments.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.