Chikungunya is a disease transmitted to humans by mosquitoes in Africa, Asia, and the Americas, with sporadic cases reported in other locations. Its symptoms often resemble those of dengue and Zika, resulting in common misdiagnoses. The illness causes fever and severe joint pain, which can be debilitating and may persist for different durations.1
The chikungunya virus (CHIKV), an alphavirus, is the causative agent of this serious illness. To facilitate viral RNA replication, CHIKV creates distinctive organelles called spherules on the plasma membrane of infected cells. While some viral protein structures within these spherules have been characterized, the full structure of the organelle is still not fully understood.
Researchers at Umea University employed Thermo Scientific Krios Cryo-TEM (cryo-transmission electron microscope) along with Thermo Scientific Amira Software to explore the structural arrangement of the spherules. Cryo-electron tomography of the plasma membrane uncovered clusters of balloon-like spherules attached to the membrane.2
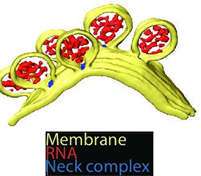
Figure1. 3D recostructure of the spherules at the plasma membrane. Yellow: plasma membrane, red: viral RNA, and blue: protein complex sitting at the spherule necks. Image Credit: Thermo Fisher Scientific - Software
As shown in the figure above, the spherules possess a circular shape with diameters between 50 and 70 nm. The tomograms also displayed a filamentous structure within the membrane buds. The location and size of these filaments suggest that they are likely viral RNA, possibly in its double-stranded RNA replicative form.
Investigating the structure of viral filaments in spherules
Researchers proposed that analyzing the visible filaments within the spherules could provide insights into the quantity of viral RNA contained in each spherule.
To facilitate this, they employed the automated filament tracing feature available in Amira Software, which allows for the tracing and quantification of genetic material in individual spherules. This tool has demonstrated effectiveness in both quantifying and analyzing filament structures.
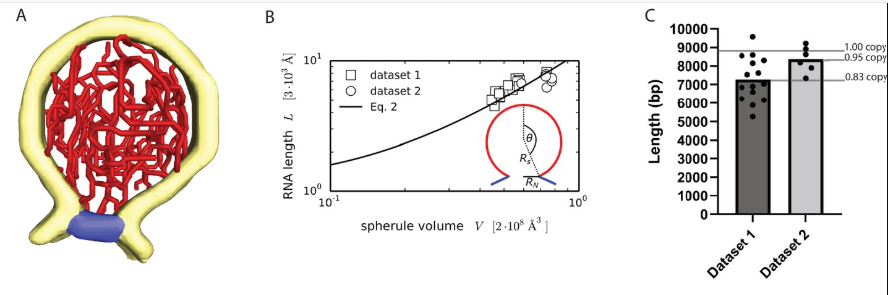
Figure 2. Characterization and quantification of the viral RNA. A: 3D structure of the ds RNA filament obtained by Amira Software. Yellow: membrane, Red: RNA and blue: neck complex base. B: Increase of RNA lengths with spherule volume shown in a graph. C: Estimation of dsRNA length (in base pairs) for two datasets, and average copy numbers per spherule. Image Credit: Thermo Fisher Scientific - Software
The total length of viral filaments within the spherules was measured, revealing an approximate length of 18,600 ± 2900 Å and 21,400 ± 1600 Å per spherule (Figure 2B), which is equivalent to about 7300 ± 1150 and 8400 ± 600 base pairs per spherule across the two datasets (Figure 2). Notably, this length represents about 80-90 % of a full-length replicon RNA, which consists of 8820 base pairs.
This finding indicates that each spherule likely contains a complete copy of the template RNA strand, with a substantial portion in a double-stranded configuration. This insight is crucial for understanding the virus's replication mechanisms and may guide future research and therapeutic approaches.2
In conclusion, the investigation of RNA filaments has provided detailed insights into their organization within spherules. In addition to characterizing the RNA filaments, the authors examined protein and lipid interactions. Their research on viral proteins identified crucial roles for the non-structural proteins (nsP1 - nsP4) in stabilizing the spherule structure and aiding RNA replication.
Meanwhile, lipid characterization underscored the significance of specific lipid compositions in membrane curvature and stability. By integrating these findings, the study offers a comprehensive understanding of the interactions among RNA, proteins, and lipids in preserving the structural integrity and functionality of viral replication organelles.
Thermo Fisher Technology
Thermo Scientific's Amira Software offers advanced visualization, segmentation, and analysis capabilities, facilitating the examination and interpretation of complex datasets.
As demonstrated in this study, Amira enabled the revelation of the 3D structure of the chikungunya spherule at the plasma membrane and allowed for quantitative analyses to estimate viral RNA.
The software's ability to handle large datasets and produce accurate 3D reconstructions and quantitative analyses significantly enhanced the research, resulting in more precise and comprehensive findings.
Amira for Cell biology webinar teaser
Watch on-demand how to maximize your cell biology images with Amira Software
References
- WHO (2022). Chikungunya. (online) Available at: https://www.who.int/news-room/fact-sheets/detail/chikungunya.
- Laurent, T., et al. (2022). Architecture of the chikungunya virus replication organelle. eLife, (online) 11, p.e83042. https://doi.org/10.7554/eLife.83042.

Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.