The precise insertion of DNA sequences using the homology-directed repair (HDR) pathway is one potential use of the CRISPR/Cas genome editing technology. Various circumstances could influence the effectiveness of this strategy, but picking the appropriate HDR template is undoubtedly crucial.
GenScript now provides high-quality, sequence-verified HDR templates to help users edit their CRISPR experiments more effectively.
CRISPR HDR knock-in templates
Accurate and minimally hazardous ssDNA GenExact™ for effective CRISPR KI
Image Credit: GenScript
- Appropriate for constructing animal models
- Suitable for T-cell therapy
Large-scale long gene knock-in using closed-end GenWand™ dsDNA.
Image Credit: GenScript
- Ideal for screening and scale up
- Ideal for long-gene CRISPR KI
GenExact™ ssDNA
- Precise knock-in, minimized off-target effect
- Research to cGMP grade
- 150–5000 nt
- High purity and sequence verified
- µg to mg scale
- Minimal cytotoxicity
GenWand™ dsDNA
- µg to g scale
- Research to cGMP grade
- More suitable for scale-up
- High purity and sequence verified
- Covalently closed ends protection for better accuracy
- 2–10 kb
Resources
Highly efficient non-viral T cell engineering process at clinical scale with GenExact™ ssDNA
Collaboration with UCSF’s (University of California San Francisco) Marson’s lab revealed that:
- At a GMP-compatible scale, the KI efficiency of GenExact™ ssDNA through electroporation without any enhancer can reach 46.2%
- GenExact™ ssDNA produced much higher KI-positive live cells than the patient’s projected dose (100*10e6). BCMA-CAR cells were effectively killed in in vitro experiments
- When compared to internally produced HDRTs (homology-directed repair templates), GenExact™ ssDNA consistently outperformed them due to its lower levels of toxicity and higher knock-in efficiency
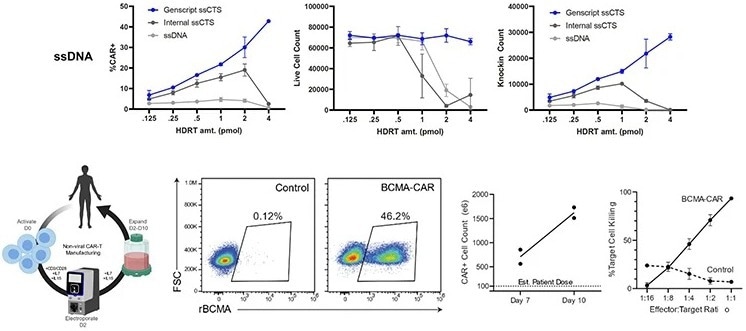
Brian R Shy. et al., bioRxiv(2021). Image Credit: GenScript
GenWand™ dsDNA outperforms PCR products in knocking in larger genes
Comparing GenWandTM dsDNA to internal PCR dsDNA (38% vs. 21%), the KI efficiency was found to be 80% higher.
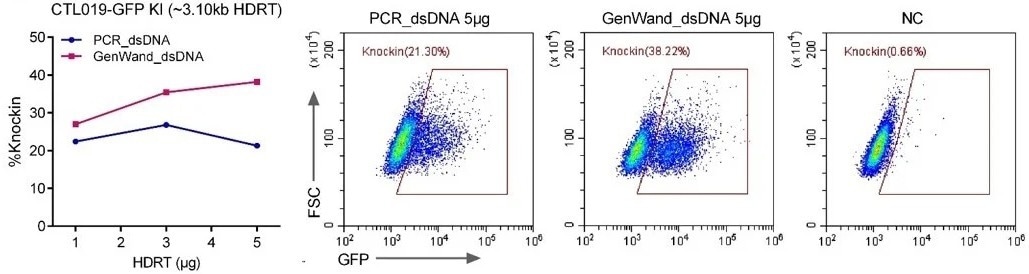
GFP KI at RAB11A Locus in HEK293 cells using electroporation with 5 ug of PCR dsDNA and GenWand™ dsDNA templates. Image Credit: GenScript