Scientists are anxiously searching for an effective antiviral to combat severe or critical COVID-19, which has already caused over a million deaths worldwide. A new study published on the preprint server bioRxiv* reports the elucidation of the mechanism of inhibition of SARS-CoV-2 by a non-nucleotide inhibitor called suramin. This is the first such compound to be shown to have such activity and shows promise for use as a repurposed drug against COVID-19.
Compared to the earlier highly pathogenic SARS-CoV and MERS-CoV, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has much greater transmissibility and has therefore caused a pandemic rather than a local outbreak. Suramin is a drug that has been used to treat African sleeping sickness and river blindness for over a hundred years. Suramin has also been shown to be an inhibitor against a wide variety of viruses, including Zika and Chikungunya viruses, indicating its diverse mechanisms of action against multiple viruses.
For instance, it can prevent viral attachment, entry, and release from host cells by its interactions with viral capsid proteins. Recent research has shown it to be an in vitro inhibitor of SARS-CoV-2 infection in cell culture by preventing viral entry. The current study shows this to be due to its potent inhibition of SARS-CoV-2 RNA-dependent RNA polymerase (RdRp), an essential viral lifecycle enzyme. Suramin is 20 or more times as powerful as remdesivir, which is the only currently approved anti-COVID-19 drug.
cryoEM studies reveal the binding of suramin to the RdRp catalytic site to prevent RNA and primer binding. This serves as a rationale to repurpose suramin for this virus as well as providing a design template of a structure that can be tweaked for newer derivatives of suramin.
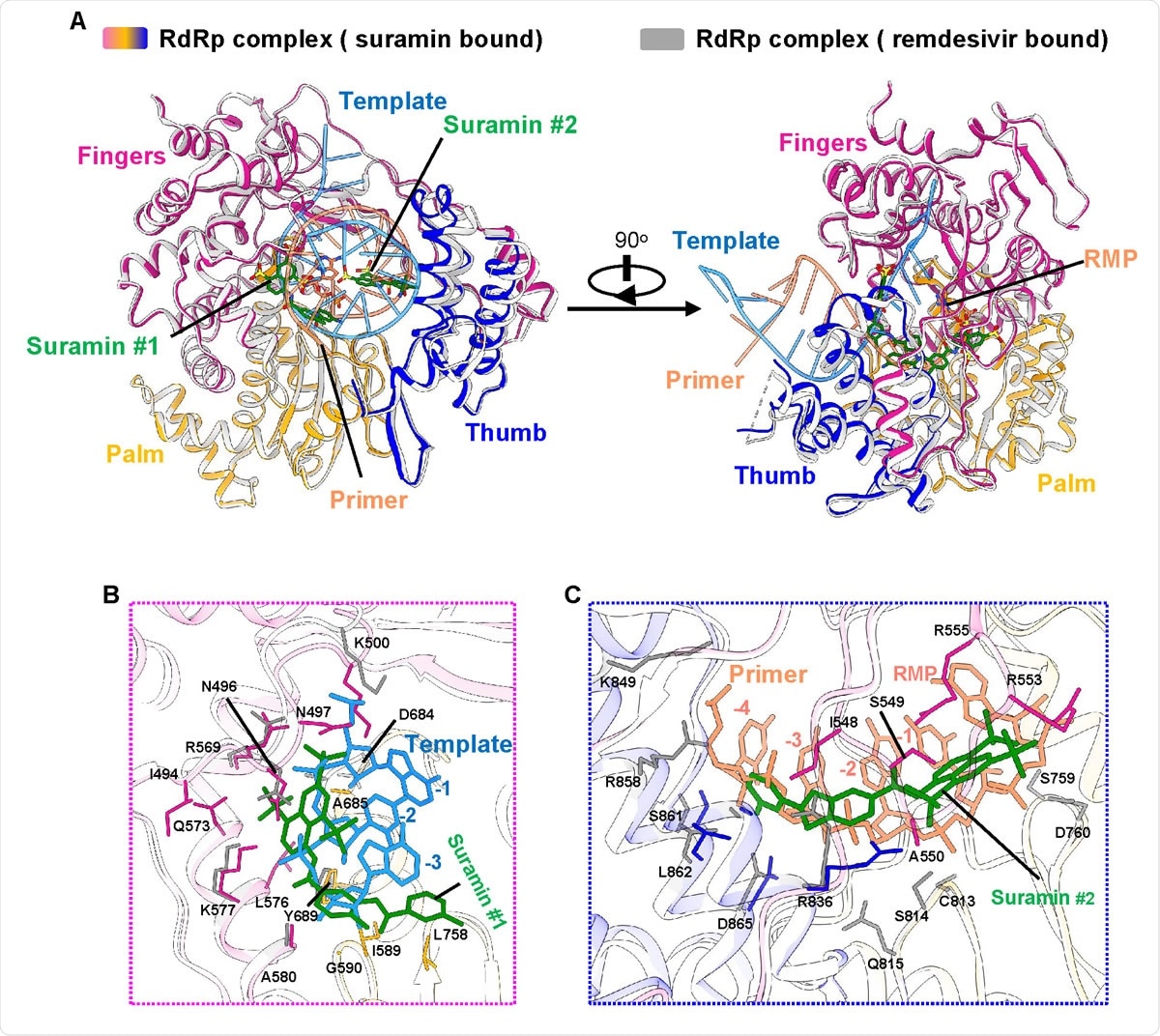
Inhibition mechanism from comparison with the remdesivir-bound RdRp structure A. The two overall views of the RdRp-suramin complex overlapped with the remdesivir-bound RdRp structure (PDB ID: 7BV2). For clarity, only the polymerase domains are presented. The remdesivir bound RdRp structure is set as light gray, the template RNA is set as cyan, and the primer RNA is set as red. B. Close view of suramin #1 overlapped with RNA template strand. C. Close view of suramin #2 overlapped with RNA primer strand.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Inhibition of RdRp by suramin
SARS-CoV-2 has a core RNA polymerase made up of non-structural protein nsp12 and two accessory subunits nsp7 and nsp8. This complex is necessary for RNA-based RNA synthesis. This is almost entirely inhibited by 8-32 μM suramin compared to 100-1000 μM of remdesivir triphosphate form (RDV-TP). The same trend was seen when it comes to the complete abolition of RdRp-RNA complex formation, which required 100 μM of suramin but >5 mM of RDV-TP. The half-maximal inhibition concentration (IC50) of suramin is 0.26 μM but 6.21 μM for RDV-TP.
This indicates that suramin is 20 times or more potent than RDV-TP. In cell culture, the former was shown to inhibit the duplication of the virus in Vero cells, the half-maximal effective concentration (EC50) being ~2.9 μM comparable to that of remdesivir.
Why does suramin show weaker effects in cell culture? Researchers speculate that this is due to the high negativity of this molecule, which reduces its uptake by host cells. The CC50 (concentrations of drug required to reduce cell viability by 50%) of suramin is over 1000 μM, indicating its high margin of safety.
The structure of the RdRp-suramin complex
Using cryo-EM, the current researchers found that the RdRp-suramin complex has a very similar structure to the apo RdRp complex. Nsp 12 has the same right-hand palm-fingers configuration with a catalytic active site comprising seven motifs (A-G) that are highly conserved between coronaviruses. Two molecules of suramin fit into the catalytic chamber, one formed by motif G and motif B.
Chemically, suramin has a two-fold symmetry with a urea linker at the center. However, only half of it is defined clearly on the EM map, lacking the urea linker. The interactions between the RdRp and suramin involve hydrogen bonds, charge interactions, and hydrophobic packing interactions.
A study of the alignment with RdRp from several viruses suggests that residues that come in contact with suramin are conserved, indicating the broad applicability of suramin to multiple viral infections.
Suramin Inhibits SARS-CoV-2 RdRp
Comparing the RdRp-suramin complex to the remdesivir-bound RdRp complex shows that the two suramin molecules occupy specific positions on the RNA strand and the primer strand, blocking the binding of the duplex RNA template-primer strand to the active site of the enzyme. Simultaneously it prevented the entry of nucleotide triphosphate into the catalytic site. This directly blocked RdRp catalytic activity.
This mechanism refers to SARS-CoV-2 RdRp inhibition, which is different from the suramin-mediated inhibition of the norovirus RdRp, though this also involves the binding of two molecules of suramin. Both suramin binding sites show half of the suramin molecule, but in comparison to the earlier enzyme, only one of the two sites shows a partial overlap. In the latter case, there is an indirect inhibition of RdRp polymerization since one binding site overlaps the proposed nucleotide entry channel.
When other suramin derivatives are used, they bind at different orientations and conformations, and may thus also inhibit the SARS-CoV-2 RdRp.
Suramin derivatives Inhibit SARS-CoV-2 RdRp
Suramin-based drugs have been tested for chemotherapy of cancers and parasitic infestations. When screened in vitro, all the eight tested compounds were efficient RdRp inhibitors with five-fold potency relative to suramin. With micromolar effective concentrations, the cytotoxicity is low, indicating their potential utility for COVID-19 treatment. However, the previously observed discrepancy in the biochemical potency and cell-based assays is still present, indicating that these compounds are not well taken up by living cells in the host. Thus, the use of glycol chitosan-based nanoparticles to carry these drug particles into the cells may enhance their bioavailability in pulmonary tissues and their potency as viral inhibitors.
Implications
The researchers show that suramin is a direct and potent viral RdRp inhibitor by direct inhibition, with structural analysis showing the mechanism of binding and inhibition. This is the first non-nucleotide inhibitor of the SARS-CoV-2 RdRp, and indicates the potential to develop suramin analogs and drug formulations as potent and effective inhibitors of viral replication.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Yin, W. et al. (2020). Structural Basis For Repurposing A 100-Years-Old Drug Suramin For Treating COVID-19. bioRxiv preprint. doi: https://doi.org/10.1101/2020.10.06.328336. https://www.biorxiv.org/content/10.1101/2020.10.06.328336v1
- Peer reviewed and published scientific report.
Yin, Wanchao, Xiaodong Luan, Zhihai Li, Ziwei Zhou, Qingxing Wang, Minqi Gao, Xiaoxi Wang, et al. 2021. “Structural Basis for Inhibition of the SARS-CoV-2 RNA Polymerase by Suramin.” Nature Structural & Molecular Biology 28 (3): 319–25. https://doi.org/10.1038/s41594-021-00570-0, https://www.nature.com/articles/s41594-021-00570-0