The coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2), is characterized by a range of clinical presentations. The vascular complications of the condition include a range of various coagulopathies that cause bleeding and thrombocytopenia or a hypercoagulable state. A new preprint research paper posted to the medRxiv* server describes the role of a fibrinogen-related protein in inducing these clinical features of COVID-19.
Earlier research has shown a change in circulating cytokines involved in the inflammatory and coagulation pathways, indicating dysregulation of these biomarkers and the endothelium. These include fibrin(ogen), D-dimer, P-selectin and von Willebrand Factor (VWF), C-reactive protein (CRP), and other cytokines that bind to endothelial receptors.
During various stages of progressive COVID-19, VWF and fibrinogen levels drop while the related D-dimer shows a rise. These are predictive of a subsequent cytokine storm which indicates systemic inflammation and a poor prognosis.
This is further exacerbated by the occurrence of numerous microclots in the lungs, as well as preformed amyloid clots in the plasma. These patients also show signs of damage to red cells and platelets.
Viral spike protein and inflammation
The viral membrane proteins are closely related to the virulence of the disease. The spike protein is the main infectivity and virulence factor. This membrane glycoprotein has two subunits, the S1 and S2.
The S1 subunit mediates attachment to the host target cells at the angiotensin-converting 2 (ACE2) surface receptors. This is followed by viral entry and productive infection, brought about by a conformational change in the S1 subunit that is triggered by spike-ACE2 binding.
The spike exists as homotrimers, protruding from the viral surface, and contains an N-terminal domain (NTD) outside the membrane, a transmembrane domain and an intracellular C-terminal domain (CTD). It is heavily glycosylated, which helps evade the host cell immune response.
The shed spike protein is detectable in many organs, and the S1 subunit has been found in the brain. The researchers suggest that free spike protein can be released from the viral membrane spontaneously. Following infection, the host cell can also release the receptor-binding domain (RBD)-containing S1 subunits.
The isolated S1 subunit as a focus of inflammation is the subject of the current paper.
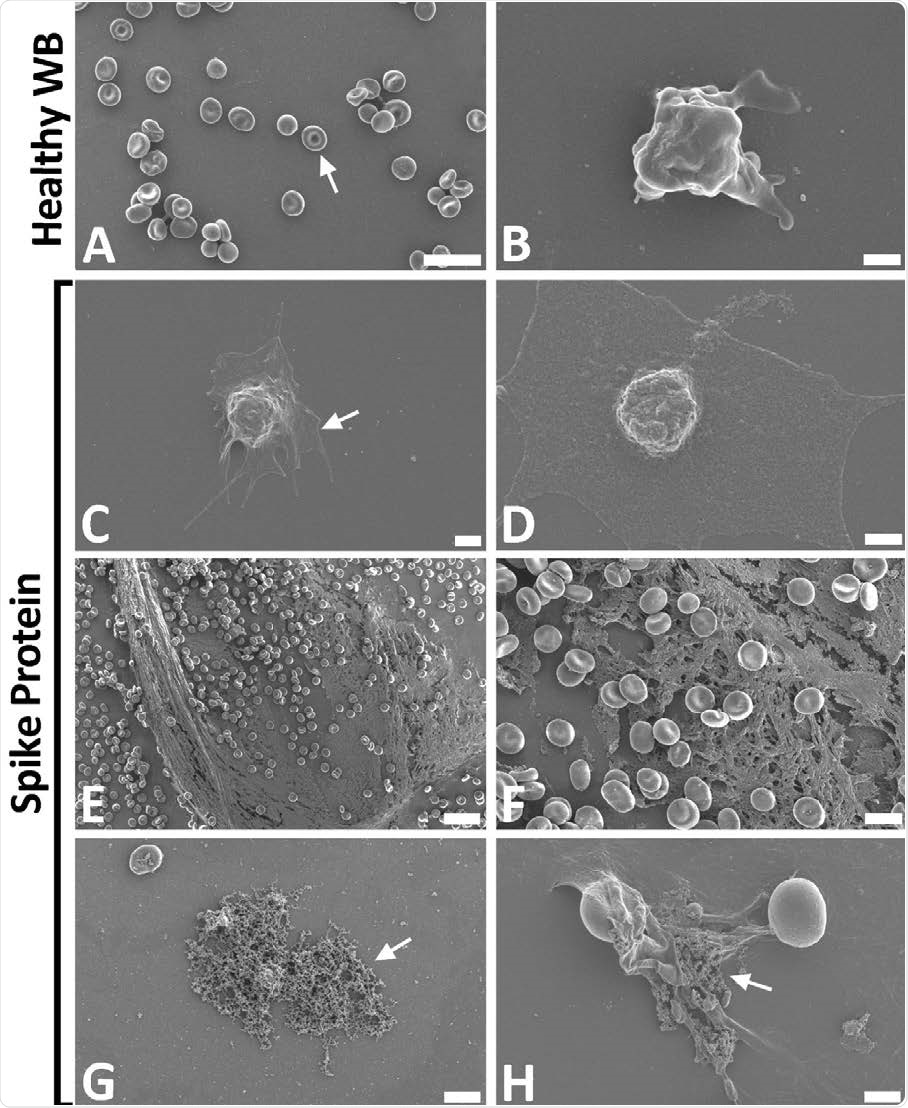
Representative scanning electron micrographs of healthy control whole blood (WB), with and without spike protein. A and B) Healthy WB smears, with arrows indicating normal erythrocyte ultrastructure. C to H) Healthy WB exposed to spike protein (1 ng.mL-1 final concentration), with C and D) indicating the activated platelets (arrow), E and F) showing the spontaneously formed fibrin network and G and H) the anomalous deposits that is amyloid in nature (arrows) (Scale bars: E: 20μm; A: 10μm; F and G: 5μm; H: 2μm; C: 1μm; B and D: 500nm).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Amyloid deposits in response to spike protein
The researchers examined the fluorescent amyloid signals in abnormal clots and in healthy platelet-poor plasma (PPP) with or without spike protein.
This showed a marked increase in dense abnormal amyloid clots, called amyloid deposits, in PPP to which spike was added, with or without thrombin. Thrombin alone also created an extensive fibrin clot. However, there was a significant increase in the percentage area of amyloid deposits.
The greatest change followed the addition of both spike and thrombin.
Platelet activation
When whole blood was exposed to spike protein even at low concentrations, the erythrocytes showed agglutination, hyperactivated platelets were seen, with membrane spreading and the formation of platelet-derived microparticles.
In all samples, spontaneous amyloid deposits formed after exposure to the spike protein without the need for thrombin exposure.
Clotting in microfluidics channels
Microfluidics systems were set up to simulate extensive endothelial damage, with resulting hypercoagulability. This showed that COVID-19 produced changes in the clotting profile of the PPP.
Clot formation in healthy PPP occurred slowly and gradually, to a moderate size, and with orderly clot layers that allowed blood flow to occur through the channel's center. These clots were easily removed by flushing the channel at 1 mL/min.
The PPP from COVID-19 patients showed large disorderly clots that often projected into the channel's center and obstructed the flow. These clots were impossible to dislodge at the earlier flow rate or even at a higher flow.
Again, large clots formed in PPP from COVID-19 patients when it was exposed to thrombin in about 90 seconds. However, most of the clotting happened in one burst, with not much propagation of the clot thereafter, indicating rapid consumption of the thrombin.
This was not the case with PPP exposed to spike protein, where a fibrous laminar clot was combined with a chaotic clot. Moderate flow disruption was also observed. These clots could also be removed with similar ease. This intermediate state could be due to the absence of multiple other biological factors that may have hindered the formation of the characteristic clots seen in COVID-19 patients.
Mass spectrometry
The results of mass spectrometry of the healthy PPP with spike protein showed changes in the structure of the beta and gamma fibrin(ogen) proteins, together with complement 3 and prothrombin. These proteins showed resistance to degradation by trypsin, a powerful proteolytic enzyme, in the presence of spike protein.
What are the implications?
The researchers show that the spike S1 not only interacts directly with both platelets and with the key clotting protein fibrinogen and its activated form, fibrin, causing changes in the protein that, in turn, alter the way blood clots.
In PPP, the addition of thrombin was found to induce fibrinogen's polymerization into a fibrin mesh. Exposure to spike protein was shown to precipitate dense clots.
When spikes and thrombin were added to healthy PPP, the formation of abnormal amyloid deposits was increased. These also showed significant changes in the blood cells' ultrastructure, including the red cells and platelets.
The presence of extensive spontaneous fibrin networks following the addition of the spike protein to whole blood matches the ultrastructural appearance seen on COVID-19-positive blood smears. Here again, the primary features were anomalous clotting, amyloid in the clots, and spontaneous fibrin network formation.
The study also shows that it may alter blood flow in COVID-19. The microfluidics simulation showed that the PPP from COVID-19 patients, which is almost pure fibrinogen, formed large obstructing clots. The PPP "may have contained downstream effects of some endothelial changes that would give rise to the hypercoagulable state that is characteristic of the disease."
"We suggest that, in part, the presence of spike protein in circulation may contribute to the hypercoagulation in COVID-19 positive patients and may cause severe impairment of fibrinolysis. Such lytic impairment may be the direct cause of the large microclots we have noted here."
Thus, the free S1 subunit has harmful effects on the host even without direct infection of the cells themselves. This strengthens the case for targeting the spike protein via antibodies and vaccines.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Grobbelaar, L. M. et al. (2021). SARS-CoV-2 spike protein S1 induces fibrin(ogen) resistant to fibrinolysis: Implications for microclot formation in COVID-19. medRxiv preprint. doi: https://doi.org/10.1101/2021.03.05.21252960. https://www.medrxiv.org/content/10.1101/2021.03.05.21252960v1
- Peer reviewed and published scientific report.
Grobbelaar, Lize M., Chantelle Venter, Mare Vlok, Malebogo Ngoepe, Gert Jacobus Laubscher, Petrus Johannes Lourens, Janami Steenkamp, Douglas B. Kell, and Etheresia Pretorius. 2021. “SARS-CoV-2 Spike Protein S1 Induces Fibrin(Ogen) Resistant to Fibrinolysis: Implications for Microclot Formation in COVID-19.” Bioscience Reports 41 (8): BSR20210611. https://doi.org/10.1042/BSR20210611. https://portlandpress.com/bioscirep/article/41/8/BSR20210611/229418/SARS-CoV-2-spike-protein-S1-induces-fibrin-ogen.