A pathogen's ability to infect or damage its host tissues is determined by the virulence factors. These are often molecules synthesized by bacteria or viruses and encoded in their genome, but they may also be acquired from the environment via transmissible genetic elements.
 Space-filling molecular model of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Scientific background. 3D illustration. Image Credit: Maryna Olyak / Shutterstock
Space-filling molecular model of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Scientific background. 3D illustration. Image Credit: Maryna Olyak / Shutterstock
Examples of virulence factors
The virulence factors of bacteria and viruses can differ drastically due to the obvious differences in their composition. A bacteria's virulence factors can be based on, for example, the capsule and flagella, which would not apply to a virus. However, a virus's virulence factors can depend on the proteins it co-opts in the host cell to synthesize those virulence factors.
Many bacterial capsules, such as macrophages and neutrophils, prevent the immune system from detecting the bacteria. The capsules' ability to evade the immune system can let the bacteria go undetected unless antibodies are developed to match the capsular antigens.
Movement and attachment are also important considerations for bacterial virulence. The flagella, which aids in movement, can help bacteria spread. The flagellum is a key virulence factor in urinary tract infections because it helps the bacteria spread up the urethra. Pili are shorter filaments that aid in attachment. Greater ability to adhere to tissues improves the bacteria's infectivity.
One of the more significant virulence factors of bacteria is exotoxins. When released by bacteria, exotoxins can interrupt and dysregulate critical cellular processes. They can also aid in the bacterial proteins' capacity to invade tissues. Similarly, viral virulence factors can include efficient replication and protein synthesis to increase virulence.
Virulence factors and treatments
Finding treatment options for pathogens is a constant struggle, especially in the age of antibiotic resistance. It is becoming increasingly common that virulence factors are identified at the molecular and genetic level, which opens up the possibility of targeting virulence factors as a form of treatment against pathogens.
Many antibiotics kill both pathogenic and commensal, not harmful, bacteria, which can lead to adverse side effects. Since commensal bacteria will not likely share a pathogenic bacteria's virulence factors, drugs targeting virulence factors can theoretically prevent many of these deleterious effects. Similarly, infections caused by or related to biofilms are not always treatable by antibiotics and could instead be treated by virulence factor-related drugs.
Many virulence factors focus on the evasion of the host immune system. Some of these, such as meningococcal sepsis and some pneumonia, can debilitate an otherwise healthy patient. While the principle of these virulence mechanisms as therapeutic targets is sound, there is not much experimental evidence of the merit of this approach.
Less specific drugs are not the only way to target virulence factors. Some virulence factors are believed to be comprised of, or heavily dependent on, the host's nutritional status both before and during infection.
Therefore, proper nutritional care of patients can prevent virulence factors from being as successful as they would otherwise be. This is especially true for diseases that commonly have a lack of appetite as a symptom, such as COVID-19.
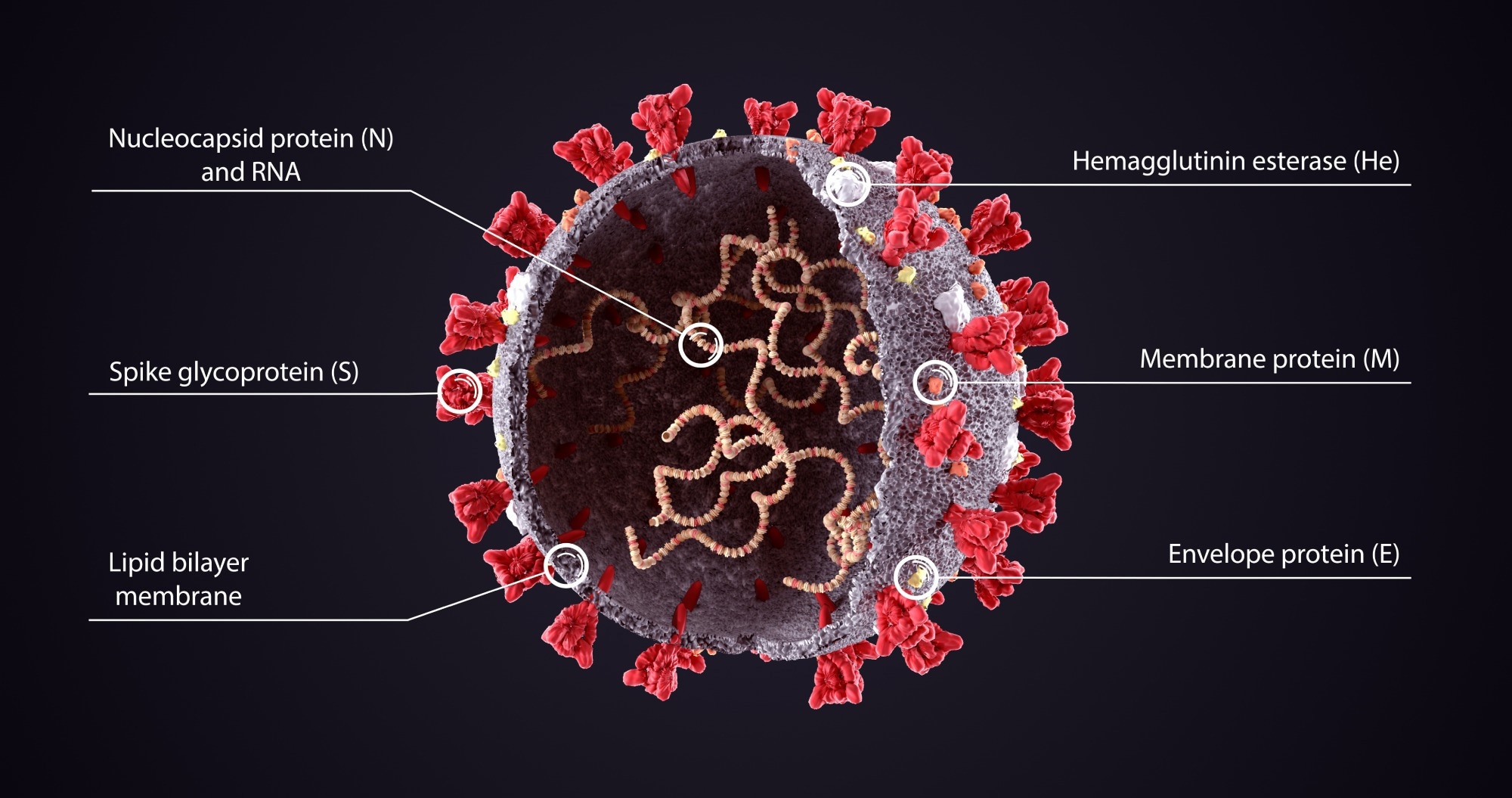 Realistic 3D Illustration of COVID-19 Virus Structure Diagram. Corona Virus SARS-CoV-2, 2019 nCoV virus. Image Credit: Orpheus FX / Shutterstock
Realistic 3D Illustration of COVID-19 Virus Structure Diagram. Corona Virus SARS-CoV-2, 2019 nCoV virus. Image Credit: Orpheus FX / Shutterstock
Virulence factors and COVID-19
COVID-19 has had huge impacts worldwide, partly due to its infectiousness. This high level of infectiousness is enabled by its virulence factors, which are being intensely studied. Examples of hypothesized virulence factors of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are the following examples.
SARS-CoV-2 unique FCS
There is an FCS containing multi-basic amino acids (PRRAR) insertion within the spike protein of SARS-CoV-2 at the S1/S2 intersection. This FCS is of evolutionary gain and is thought to be involved in its high infectivity and transmissibility.
Manipulation of host immune response
SARS-CoV-2 is thought to have virulence factors directed against host immune responses. Current data suggests that the SARS-CoV-2 tricks the host into a delayed hyperactive innate immune response, leading to the cytokine storm commonly seen in severe COVID-19 cases. This reduces T cell response.
Spike proteins
The SARS-CoV-2 spike proteins facilitate entry into host cells. The spike protein enables the fusing of the host and viral cell membranes, initiating virus infection. Reports also suggest that the spike protein's S2 subunit is adapted to protect the virus from the host immune system through its N-linked glycans.
Mimicking of an epithelial ion channel
A proposed molecular mechanism of SARS-CoV-2 suggests that once a virus has entered the cell, it competes for FURIN with a structurally similar human epithelial sodium channel α-subunit (ENaC- α). Some literature proposes that ENaC- α has a vital role in immune cell activation and cytokine-mediated ARDS. Hence, this mechanism could be key to the arising of ARDS in severe COVID-19.
Driving of host cell molecular pathways
Studies have shown that SARS-CoV-2 proteins are phosphorylated by the host proteome, initiating host cell kinase activation and growth factor receptor signaling. This essentially results in the host protein machinery being hijacked by the virus.
Summary
Understanding SARS-CoV-2 and its virulence factors provides key information about COVID-19 pathogenesis, its associated complications, and why it is so transmissible compared to previous SARS viruses; targeting these virulence factors could be key to therapeutic measures for COVID-19, a subject being explored by many studies currently.
Bacterial virulence factors an introduction
References
- Briguglio, M., Pregliasco, F., Lombardi, G., Perazzo, P. and Banfi, G., 2020. The Malnutritional Status of the Host as a Virulence Factor for New Coronavirus SARS-CoV-2. Frontiers in Medicine, 7.
- Cross, A., 2008. What is a virulence factor? Critical Care, 12(6), p. 197.
- Kumar, A. et al. (2020). SARS‐CoV‐2‐specific virulence factors in COVID‐19. Journal of Medical Virology.
- Webb, S., and Kahler, C., 2008. Bench-to-bedside review: Bacterial virulence and subversion of host defenses. Critical Care, 12(6), p. 234.
Further Reading
Last Updated: Feb 28, 2024