Cytosine rich DNA sequences can fold into a structure called an i-motif. These structures typically appear in promoter regions on DNA, and are thought to help with gene regulation.
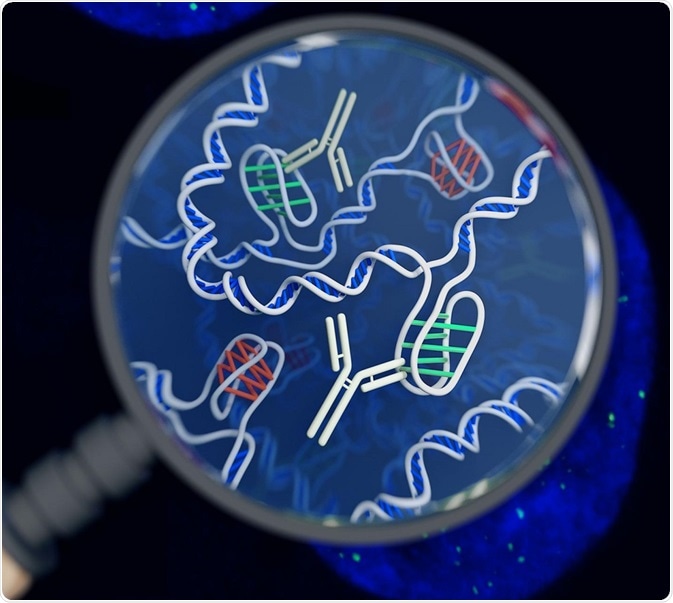 An artist's impression of the i-motif DNA structure inside cells, along with the antibody-based tool used to detect it. Image Credit: Chris Hammang / Shutterstock
An artist's impression of the i-motif DNA structure inside cells, along with the antibody-based tool used to detect it. Image Credit: Chris Hammang / Shutterstock
Around 98% of the DNA consists of non-coding regions, which are often involved in transcriptional and translational regulation of the genome. These regions consist of repetitive sequences.
B-DNA, or the right-handed double helix, is the most prevalent DNA structure under normal physiological conditions. But under specific conditions, the DNA may fold into hairpin or Z DNA structures where the DNA has a left-handed twist with a zigzag sugar phosphate backbone.
Non-B-DNA structures may lead to genomic instability and disorders. The G-rich sequences can fold in to a non-B-DNA structure called G-quadruplex. These structures have been found in vivo also as they are stable at physiological temperature and pH.
Similarly, C-rich regions can fold in to a structure called i-motif or i-tetraplex or i-DNA. These are four stranded DNA structures which are held together through the intercalation of cytosine base pairs.
Structure of the i-motif
The i-motif is a tetrameric or four stranded structure consisting of two parallel duplexes (double strands) with the sequence d(TCCCCC). The two duplexes combine in an anti-parallel manner in the i-motif. This combination occurs by intercalation or insertion of cytosine-cytosine base pairs.
For the structure to form, one of the cytosine in the base pairs must be protonated and the other must b. C-C base pairs are bonded by three hydrogen bonds, which form stronger bonds than conventional G-C pairs. The base pair energy of C-C bond in an i motif is 169.7 kJ/mol, while the base pair energy of Watson Crick G-C base pair is 96.6 kJ/mol.
The structure of i-motif was found using NMR, where it was found that the length of cytosine sequences ranged from 3 to 12 bases, and it may also have thymine residues in between the cytosine sequences. The intercalation in i-motifs can occur in different ways leading to formation of two form;s R- and S-forms.
The name ‘i-motif’ was chosed as it is the only nucleic acid structure with intercalated base pairs. Although there is a lack of stacking interactions between the consecutive base pairs, the intermolecular C-H··O hydrogen bonding network between the deoxyribose sugar of antiparallel backbone stabilizes the structure.
The protonation of cytosine reduces the negative charge of the backbone and facilitates the formation of the fur-stranded structure. The base pair distance is 3.1 Å which is similar to A-DNA (2.1 Å). The helical twist between the C-C base pair is smaller than B-DNA (i motif: 12−16°, B-DNA: 36°).
Stability of the i-motif
The stability of a structure is usually determined by its melting point, or Tm, which is temperature at which a folded structure transitions to an unfolded state. This can be induced by heating the DNA sample. This transition is measured using molecular absorption or circular dichroism techniques and is known as the temperature of melting (Tm).
The Tm of a DNA sample is dictated by its nucleotide sequence and ionic strength. As one of the C- bases is protonated, the pH of the medium plays a critical role in the stability of i-DNA. At pH 4-7, the bases are partially protonated and i-structures form.
If the pH increases, the C bases undergo deprotonation and the structure unravels. If the pH is too low, all the C-bases are protonated and thus fail to form the four stranded structure. This is in contrast to the Watson and Crick base pairs, whose stability is not dependent on the pH of the medium.
The number of C-C base pairs also determines the stability of the i-motifs. The presence of six or less C bases lead to intermolecular folding of i-motifs, whereas longer than six C residues promote the formation of intramolecular folding of i-motifs.
i-DNA versus Watson Crick structures
Studies have investigated the relative presence of tetraplex structures (i-motifs and G-quadraplex) and Watson-Crick duplexes. It was found that at physiological pH, the duplex structures dominated, whereas at pH lower than 5, the tetraplex structures were predominant.
The concentration of tetraplex structures at pH 7 and 25°C was less than 10% suggesting the dominance of duplex structures in vivo in physiological conditions.
In vivo presence of i-DNA
Although the Watson Crick base pairs are more stable in physiological conditions, studies have investigated the presence of i-DNA in vivo. One of the possibilities is that during transcription and replication, DNA undergoes negative supercoiling which could promote the formation of i-motifs.
Under different conditions, the physiological pH of a cell is altered, potentially promoting the formation of i-motifs. For example, cancers are associated with lower intracellular pH (6.7–7.1). Also, certain cellular processes can lead to temporary acidification of the cell which may also promote transient formation of these structures. Recently using antibodies which specifically recognize i-motifs in DNA, researchers have identified i-motifs in nuclei and regulatory regions, including promoters and telomeres.
Further Reading