Skip to:
Some decades after the first ever genetic sequence of a living organism was shared, public databases contain several hundred complete bacterial genomes. Nonetheless, only a small fraction of these species of bacteria have genomes that have been fully sequenced.
The conclusion from the study revealing this, is that each of the bacterial species, in theory, will never be fully described as new genes will be continuously added to the genome of the species with each new genomic sequence. Hence, the best way to describe a bacterial species is to take a look at the concept of the pan-genome: comprised of the all of the dispensable and core genomes of a species.
What is the Pan-Genome?
The pan-genome characterizes all of the genetic variations and singular genes within one single species. This original concept was brought to light by Tettelin et al., when six strains of Streptococcus agalactiae were sequenced.
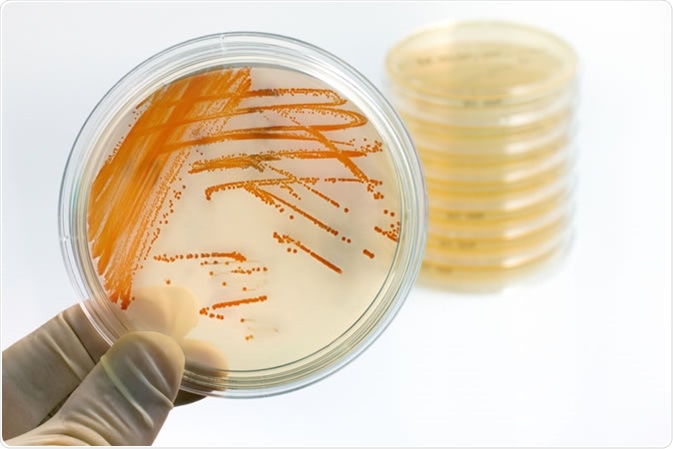
Colonies of bacteria Streptococcus agalactiae in culture medium plate. Image Credit: Angellodeco / Shutterstock
The DNS sequences obtained could be construed as a core genome (shared by all S. agalactiae isolates), which accountis for around 80% of the single genome, as well as an expendable genome consisting of strain-specific and partially-shared genes. Estimates from this study suggest that the genetic reservoir within the S. agalactiae pan-genome is immense, and that these new genes could continue to be further identified.
Examples of Pan-Genome Analysis
Lactobacillus paracasei is part of both the animal and human gut microbiome, and is therefore used in the food industry in probiotic products, or as started cultures for other dairy products. With the increase in the use of high-throughput and low-cost DNA sequencing methods it has become feasible to sequence a variety of different strains of a single species, to then determine its pan-genome. In a 2013 study, the genomes of 34 separate strains of L. paracasei were sequenced, and genomics analysis comparing each strain were executed.
Genome content and synteny were analysed, with a focus on the pan-genome. Each of the genomes were found to contain approximately 2,800 to 3,100 genes, as well as the comparative analysis identifying well over 4,200 ortholog DNA groups comprising the pan-genome of this particular species. Around 1800 of the ortholog groups comprise the protected genome core. A variety of factors that used to be linked to host-microbe biochemical interactions, such as cell-envelope proteinase, pilli, and hydrolases p75 and p40. The capability to make short branched-chain fatty acids were all found to be present in the L. paracasei core genome, found in all of the chosen strains.
The variome (the part of the genome that can vary between organisms), was found to consist mainly of hypothetical phages, proteins, transposon/conjugative elements, plasmids, and familiar functions (for example, sugar metabolism and CRISPR-associated proteins). Large variability and variety of sugar-utilization genetic cassettes were pinpointed, with each of the strains having between 25 and 53 individual cassettes, which reflect the adaptability of L. paracasei to varied niches. A phylogenomic tree was then constructed, which was based on the total genome contents together with a final analysis of any horizontal gene transfer events. It was concluded that the adaptation of these strains of L. paracasei is a complex process.
Another study also found that the pan-genome could be used as a novel tool to redefine these pathogenic species of bacteria. This was then applied to Escherichia coli and Shigella bacteria species, which have recently been the subject of some controversy in regards to their pathogenic and taxonomic positions.
After choosing specific strains that are of interest, they then selected an experimental technique: such as a bioinformatics-based method or a microarray. The bioinformatics technique offers tools serving dedicated and general purposes. Using these analyses, the study of pan-genomics can give different types of data, and then increase the understanding and knowledge of a bacterial species.
By grouping together this genomic data, it could be possible to later redefine species, to classify them built upon on their variome genomic content. In a situation where an “infinite pan-genome” exists, such as in E. coli, or Prochlorococcus marinus, could re-classification be executed yet? Definitions of “species” were often decided upon using old methods and tools. Additionally, some species are non-homogeneous by nature. Therefore, redefining how a bacterial species is determined by analysing the pan-genome may be something that can be done in the near future.
Further Reading
Last Updated: Oct 15, 2019