A team led by Purdue University researchers is the first to determine the structure of the Zika virus, which reveals insights critical to the development of effective antiviral treatments and vaccines.
The team also identified regions within the Zika virus structure where it differs from other flaviviruses, the family of viruses to which Zika belongs that includes dengue, West Nile, yellow fever, Japanese encephalitis and tick-borne encephalitic viruses.
A paper detailing the findings was published Thursday (March 31) in the journal Science and is available online.
Any regions within the virus structure unique to Zika have the potential to explain differences in how a virus is transmitted and how it manifests as a disease, said Richard Kuhn, director of the Purdue Institute for Inflammation, Immunology and Infectious Diseases (PI4D) who led the research team with Michael Rossmann, Purdue’s Hanley Distinguished Professor of Biological Sciences.
“The structure of the virus provides a map that shows potential regions of the virus that could be targeted by a therapeutic treatment, used to create an effective vaccine or to improve our ability to diagnose and distinguish Zika infection from that of other related viruses,” said Kuhn, who also is head of Purdue’s Department of Biological Sciences. “Determining the structure greatly advances our understanding of Zika - a virus about which little is known. It illuminates the most promising areas for further testing and research to combat infection.”
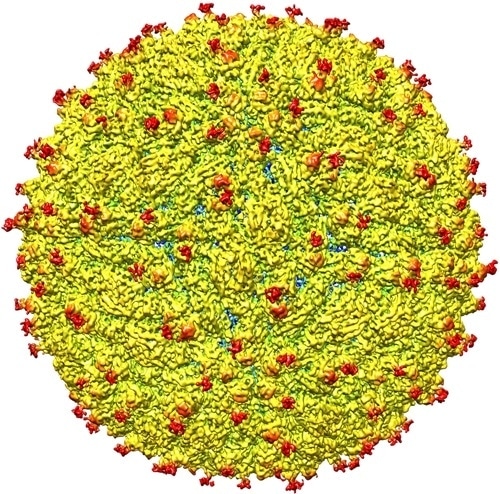
A representation of the surface of the Zika virus is shown. A team led by Purdue University researchers is the first to determine the structure of the Zika virus, which reveals insights critical to the development of effective antiviral treatments and vaccines. (Purdue University image/courtesy of Kuhn and Rossmann research groups)
The Zika virus, a mosquito-borne disease, has recently been associated with a birth defect called microcephaly that causes brain damage and an abnormally small head in babies born to mothers infected during pregnancy. It also has been associated with the autoimmune disease Guillain-Barré syndrome, which can lead to temporary paralysis. In the majority of infected individuals symptoms are mild and include fever, skin rashes and flulike illness, according to the World Health Organization.
Zika virus transmission has been reported in 33 countries. Of the countries where Zika virus is circulating 12 have reported an increased incidence of Guillain-Barré syndrome, and Brazil and French Polynesia have reported an increase in microcephaly, according to WHO. In February WHO declared the Zika virus to be “a public health emergency of international concern.”
“This breakthrough illustrates not only the importance of basic research to the betterment of human health, but also its nimbleness in quickly addressing a pressing global concern,” said Purdue President Mitch Daniels. “This talented team of researchers solved a very difficult puzzle in a remarkably short period of time, and have provided those working on developing vaccines and treatments to stop this virus a map to guide their way.”
Rossmann and Kuhn collaborated with Theodore Pierson, chief of the viral pathogenesis section of the Laboratory of Viral Diseases at the National Institutes of Health National Institute of Allergy and Infectious Diseases. Additional research team members include Purdue graduate student Devika Sirohi and postdoctoral research associates Zhenguo Chen, Lei Sun and Thomas Klose.
The team’s paper marks the first published success of the new Purdue Institute for Inflammation, Immunology and Infectious Diseases in Purdue’s Discovery Park.
The university’s recently announced $250 million investment in the life sciences funded the purchase of advanced equipment that allowed the team to do in a couple of months what otherwise would have taken years, Rossmann said.
“We were able to determine through cryo-electron microscopy the virus structure at a resolution that previously would only have been possible through X-ray crystallography,” he said. “Since the 1950s X-ray crystallography has been the standard method for determining the structure of viruses, but it requires a relatively large amount of virus, which isn’t always available; it can be very difficult to do, especially for viruses like Zika that have a lipid membrane and don’t organize accurately in a crystal; and it takes a long time. Now, we can do it through electron microscopy and view the virus in a more native state. This was unthinkable only a few years ago.”
The team studied a strain of Zika virus isolated from a patient infected during the French Polynesia epidemic and determined the structure to 3.8Å. At this near-atomic resolution key features of the virus structure can be seen and groups of atoms that form specific chemical entities, such as those that represent one of 20 naturally occurring amino acids, can be recognized, Rossmann said.
The team found the structure to be very similar to that of other flaviviruses with an RNA genome surrounded by a lipid, or fatty, membrane inside an icosahedral protein shell.
The strong similarity with other flaviviruses was not surprising and is perhaps reassuring in terms of vaccine development already underway, but the subtle structural differences are possibly key, Sirohi said.
“Most viruses don’t invade the nervous system or the developing fetus due to blood-brain and placental barriers, but the association with improper brain development in fetuses suggest Zika does,” Sirohi said. “It is not clear how Zika gains access to these cells and infects them, but these areas of structural difference may be involved. These unique areas may be crucial and warrant further investigation.”
The team found that all of the known flavivirus structures differ in the amino acids that surround a glycosylation site in the virus shell. The shell is made up of 180 copies of two different proteins. These, like all proteins, are long chains of amino acids folded into particular structures to create a protein molecule, Rossmann said.
The glycosylation site where Zika virus differs from other flaviviruses protrudes from the surface of the virus. A carbohydrate molecule consisting of various sugars is attached to the viral protein surface at this site.
Zika Animation
In many other viruses it has been shown that as the virus projects a glycosylation site outward, an attachment receptor molecule on the surface of a human cell recognizes the sugars and binds to them, Kuhn said.
The virus is like a menacing stranger luring an unsuspecting victim with the offer of sweet candy. The human cell gladly reaches out for the treat and then is caught by the virus, which, once attached, may initiate infection of that cell.
The glycosylation site and surrounding residues on Zika virus may also be involved in attachment to human cells, and the differences in the amino acids between different flaviviruses could signify differences in the kinds of molecules to which the virus can attach and the different human cells it can infect, Rossmann said.
“If this site functions as it does in dengue and is involved in attachment to human cells, it could be a good spot to target an antiviral compound,” Rossmann said. “If this is the case, perhaps an inhibitor could be designed to block this function and keep the virus from attaching to and infecting human cells.”
The team plans to pursue further testing to evaluate the different regions as targets for treatment and to develop potential therapeutic molecules, Kuhn said.
Kuhn and Rossmann have studied flaviviruses, the family of viruses to which Zika belongs, for more than 14 years. They were the first to map the structure of any flavivirus when they determined the dengue virus structure in 2002. In 2003 they were first to determine the structure of West Nile virus and now they are the first to do so with the Zika virus.
The National Institutes of Health National Institute of Allergy and Infectious Diseases funded the research through an existing grant to Rossmann and Kuhn together with colleagues Michael Diamond and Daved Fremont at Washington University, and an emergency supplement to support Zika virus research issued in February.
Purdue’s investment in the life sciences is coupled with the university’s new Pillars of Excellence in the Life Sciences Initiative. The Purdue Institute for Inflammation, Immunology and Infectious Diseases (PI4D) along with the Center for Integrative Neuroscience were two “pillars” selected to amplify Purdue’s existing strengths and complement the Purdue Moves initiatives in order to enable research at the highest level and with the greatest impact on human lives.
The initiative was implemented as a partnership of pillar leadership teams and the offices of Purdue’s Executive Vice President for Research and Partnerships and Provost.
Purdue Moves is an initiative designed to broaden the university's global impact and enhance educational opportunities for its students. All of the moves fall into four broad categories: science, technology, engineering and math (STEM) leadership; world-changing research; transformative education; and affordability and accessibility.