Today, using anatomic pathology, the differences in appearance of a normal or a malignant lesion can be difficult to tell when just using light microscopy.
How does proteomic mass spectrometry imaging (MSI) work and why could it be helpful in the differential diagnosis of malignant melanoma?
For many years, mass spectrometry has been the gold standard for identifying proteins in cells or biofluids. The breakthrough of mass spec imaging means that you can actually display these individual proteins as images in the context of the tissue section. This unites the fields of mass spectrometry and anatomic pathology.
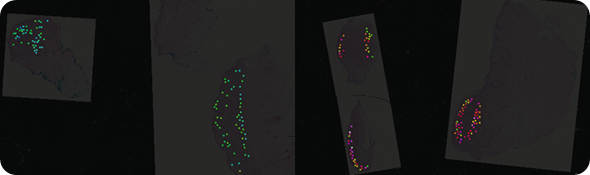
Mass spec image of a peptide at mass 1198.7. The peptide is more highly expressed in malignant lesions (red, pink, white) than in benign lesions (blue, green).
Instead of a pathologist just being dependent upon the physical appearance of the cells, they now have the additional advantage of molecular information provided by mass spec imaging, which they can use to augment the traditional examination of the cell sample.
Specifically the way we used this to develop a test was to use what's called a machine learning algorithm where the data generated is used to detect and distinguish certain molecular fingerprints between benign and cancerous tissue.
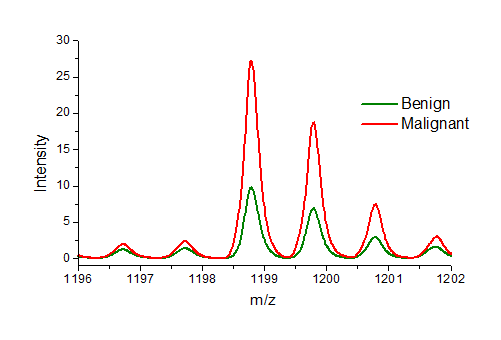
Average spectra from benign (green) and malignant (red) lesions. The peptide at mass 1198.7 is about 3 times more abundant in malignant lesions than benign.
The traditional biomarker approach of finding a protein that's specific to cancer is challenging at best because of the diversity of protein expression in cancer cells, but by generating the data in this way and letting the machine tell us what's specific to the cancer cell, we have been able to produce a publication that indicated very high levels of sensitivity and specificity.
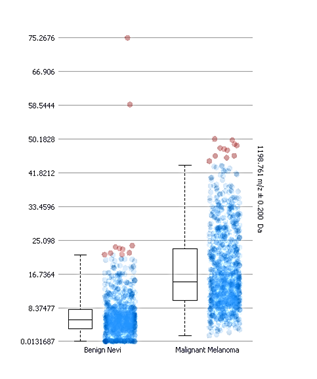
Box and whisker plots of the intensity distributions of the peptide at mass 1198.7 in benign and malignant lesions. The boxes do not overlap indicating a significant difference.
We're looking at all the subtypes of melanocytic skin lesions. There are many subtypes of skin disorders between malignant melanoma and benign pigmented nevi. In time, we'll have a database of protein expression for each type that can provide highly specific input to the pathologist.
At what stage of development is the test currently at?
We have published a proof of concept clinical study of about 85 samples, divided into a training and validation set. That was published at the American Society of Dermatopathology annual meeting last fall, in San Francisco. We are now accruing samples and conducting a follow-on, larger validation study, to look at additional subtypes of skin lesions, to do a broader study.
How sensitive and specific has the new method been shown to be?
The training set was 100% sensitive and 99% specific in its ability to differentiate melanoma from benign nevi.
What further research is needed before this test can be used in a clinical setting? What feedback have you received so far?
We've received very positive feedback from the field because melanomas can be difficult to call. A significant portion of biopsies are very challenging and, of course, a cancer cell is a cancer cell because of the things that happen at the molecular level. It makes sense that direct molecular information is going to make diagnosis more specific.
We will need to organize and put in place a CLIA diagnostic laboratory and conduct additional validation studies, at which point we can offer the tests. The beauty of the technology is that there's very little sample preparation. It's a mostly automated procedure, so that lends itself to scaling-up.
What impact do you think the test will have on detecting malignant melanoma?
I think it will be adopted. Assuming our clinical validation studies are positive and consistent with early results, then it will be a useful adjunct to the anatomic pathology that's currently in place, providing molecular information that's in the context of the tissue architecture.
It's very compelling to provide additional molecular data , as opposed to trying to change the way it's currently done. It's a good model and therefore I think it'll have a strong impact, particularly on the indeterminate biopsies.
Could this technology lead to the discovery of other clinically useful protein biomarker panels for the differential diagnosis of other cancer types?
The answer is yes. We are working on a parallel path with stage one lung adenocarcinoma. We are working with researchers at Memorial Sloane Kettering and we have plans to develop other projects.
We see the niche for the technology as being the difficult and ambiguous diagnoses where you're dealing with early stage cancer which may or may not become a malignant disease or a benign or hyperplastic condition. Those are the difficult calls that pathologists have and I think that's where we can add value with this technology.
What do you think the future holds for mass spectrometry imaging and how do Protea plan to develop the technology?
Well Protea's unique in offering mass spec imaging services. The potential for this is just breathtaking because mass spectrometry is precise analytical chemistry. It's the gold standard for identifying molecules, but the data sets from mass spec often require analytical chemists and a lot of sample preparation so, in a way, there's a detachment from the molecular data provided and how biologists would really like to see it.
Mass spec imaging ties mass spectrometry together with the world of biology, providing datasets which can be displayed in the context of the cell or tissue architecture..
It integrates anatomic pathology and analytical chemistry in a very useful and meaningful way. I think it will impact and integrate into many areas of anatomic pathology and microscopy in general.
Where can readers find more information?
https://proteabio.com/imaging
About Stephen Turner
Stephen Turner is Chief Executive Officer and Chairman of the Board, positions he has held since founding the company in July, 2001. From 1999 to 2001 he served as President and CEO of Quorum Sciences, Inc. From 1984 to 1997 he was President and CEO of Oncor, Inc.
He founded Bethesda Research Laboratories, Inc. in 1975 and served as its Chairman and CEO from 1975 to 1983, at which time BRL became the molecular biology division of Life Technologies, Inc.
Prior to commencing his career in biotechnology, Mr. Turner held the position of Director of Marketing for the Clinical Microbiology Division of Becton, Dickinson & Co. He received his B.A. from Stanford University in 1967. In 1994 he received the Ernst & Young Entrepreneur of the Year Award in Life Sciences for the Washington D.C. Region.