Neutron crystallography allows us to determine the three-dimensional structures of biological macromolecules, such as proteins, by means of the diffraction of neutrons from the regularly spaced atoms of a crystal.
A key difference between X-rays and neutrons is the way they interact with the atoms; X-rays are scattered by the electrons, whereas neutrons are scattered from the atomic nuclei. For X-rays, the strength of scattering of an element is proportional to its number of electrons, while for neutrons the scattering strength shows no correlation to the number of electrons, but rather depends on the specific nuclear forces.
The neutron image-plate detector of LADI-III at the Institut Laue-Langevin. © Institut Laue-Langevin / B. Lehn
As such, while the X-ray scattering from hydrogen, with only one electron, is very weak, with neutrons the scattering from hydrogen is of a similar magnitude to the other common elements of biological macromolecules - carbon, nitrogen, oxygen, sulphur and phosphorous.
This means that neutron crystallography can be used to locate the positions and movement of hydrogen more effectively than with X-rays, and this is important since hydrogen atoms and protons (H+) play key roles in many biochemical processes.
Another important difference between X-rays and neutrons is that since X-rays are ionizing radiation, they can cause damage to the crystals, leading to alterations to the crystal structure being determined.
To minimize these radiation damage effects, X-ray data are generally collected at cryogenic temperatures, typically 100K. In contrast, neutrons are non-destructive so data can be collected at room temperature, closer to physiological temperatures and resulting in the determination of ‘damage-free’ structures.

The neutron image-plate detector of LADI-III at the Institut Laue-Langevin. © Institut Laue-Langevin / R. Cubitt
What are the main limitations of X-ray crystallography in terms of determining the structures of protein/drug complexes?
The main limitation of X-ray crystallography when studying protein/drug complexes is the lack of information that can be gained for hydrogen.
Since proteins bind to their drug targets through directional hydrogen-bonding and non-directional hydrophobic and electrostatic interactions, knowledge of the location of hydrogen, can be key towards improving structure based drug design.
In order to ‘see’ hydrogen atoms with X-rays, extremely high-resolution data (better than 1.2Å resolution) are required - and even then only those hydrogen atoms that are non-mobile can be observed.
Mobile or highly polarized hydrogen atoms cannot be seen with X-rays, and since protons have no electrons they cannot be observed either. Neutron crystallography represents the only method by which both hydrogen atoms and protons can be visualized and in damage-free structures.
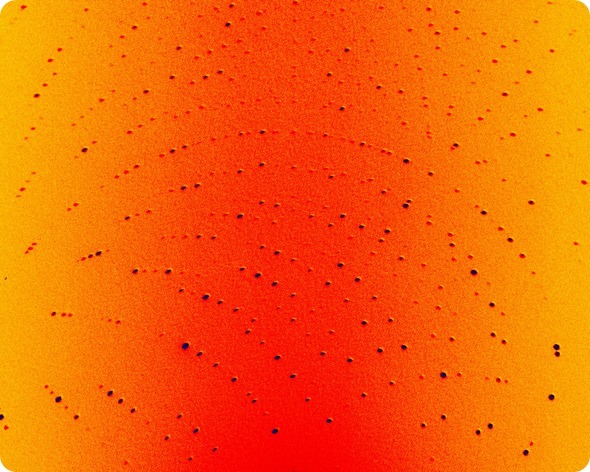
This is an example of neutron diffraction patterns from protein crystals using the LADI-III diffractometer. © Institut Laue-Langevin / M. P. Blakeley
How important is knowledge of the location of hydrogen when developing drugs?
Locating the positions of hydrogen atoms and protons using neutron crystallography allows us to observe how a drug binds to its target, providing us with information such as which hydrogen bonds are the strongest and most critical for the binding, and which hydrogen bond interactions could be enhanced to improve the binding efficiency.
Using this information we can then suggest ways to alter the molecular structure of the drug to enhance its performance and potentially reduce dosages.
Why is neutron crystallography not more widely used in structural biology?
The main challenge for neutron crystallography is that even the most intense neutron sources, such as the Institut Laue-Langevin, are many orders of magnitude less intense than synchrotron X-ray sources and so we need to compensate for this by using much bigger crystals and by collecting for longer times.
This, until recently, had limited the use of neutron crystallography as crystals of several cubic millimetres were required and data collections could take several months.
In the last few years however, we have improved the instrumentation for collecting neutron data and have reduced the sample volumes required down to around a tenth of a cubic millimetre. And instead of having to collect data for months, we can now do so in a few days to a week.
This has led to a wider range of studies becoming feasible and so we are now able to address more biologically interesting problems. Currently around 100 structures of biological macromolecules have been determined using neutron crystallography – although this is a tiny number compared to the ~100,000 structures that have been determined with X-rays, more than two-thirds of the neutron structures have been determined in the last 5 years.
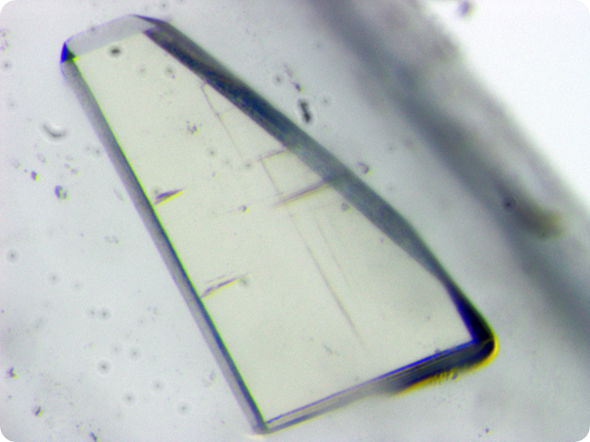
One of the HIV-1 protease crystals used for neutron data collection using the LADI-III diffractometer at the Institut Laue-Langevin. © O. Gerlits, A. Y. Kovelevsky & M. P. Blakeley.
Can you please give an overview of your recent research using neutron crystallography to probe the structure of HIV-1 protease in complex with clinical inhibitors?
This is a collaboration mainly between scientists from Georgia State University and Oak Ridge National Laboratory in the USA and the Institut Laue-Langevin in France. We are studying HIV-1 protease, an enzyme that is essential for the life cycle of HIV, since it breaks down viral polypeptides to create the proteins needed for the maturation and production of new infectious virus particles.
If HIV-1 protease’s function is blocked, HIV virus particles remain non-infectious, and so this makes the enzyme an important drug target. As such, there have been many X-ray crystallography studies of HIV-1 protease over the last twenty years or so, with the aim of designing drugs to efficiently inhibit the enzyme.
Early protease inhibitors suffered from high susceptibility to drug resistance, whereby taking the drugs led to mutant variants of HIV-1 protease being produced that bind the inhibitors much less tightly, thereby dramatically reducing their efficacy. Therefore, the second-generation protease inhibitors we have today, such as darunavir, have been specifically designed to be more resistant against mutations.
Darunavir’s structure allows it to create more hydrogen bonds with the enzyme active site, and in particular, more main-chain atoms, since the backbone of HIV-1 protease maintains its spatial conformation in the presence of mutations, meaning the hydrogen bond interactions are less likely to be disrupted. Darunavir is therefore an excellent therapy target to refine and enhance HIV treatment.
Using neutron crystallography we have determined the structures of HIV-1 protease with darunavir bound at two different pHs, providing information about the positions of all the hydrogen atoms in the enzyme/drug complex. We could observe all the hydrogen bond interactions between the drug and the enzyme, and have identified which hydrogen bonds are the strongest, and hence most critical for binding.
We also identified those interactions that could be strengthened, along with ways to potentially introduce new interactions to increase the binding affinity and reduce resistance.
How sensitive is the enzyme’s catalytic activity to pH?
At higher pH the enzyme’s catalytic activity is reduced and the structures of the enzyme/drug complex we determined at pH 4.3 and pH 6 helped us explain why this is the case.
In the low pH structure the positions of the protons between the catalytic aspartic acid residues and the hydroxyl group of darunavir are in the configuration deemed necessary for the catalytic mechanism to proceed, while in the higher pH structure, the two protons had shifted and were located in non-optimal positions.
We further analysed the structures determined at different pHs and identified differences in protonation states of residues distant from the active-site. In our recent Angewandte Chemie publication [1], we suggest that the changes in protonation states of residues distant from the active-site trigger, through electrostatic effects, the two-proton transfer in the active site.
We also performed QM/MM calculations, which support this hypothesis. I found this to be a very interesting finding and one which demonstrates the large effect changes in protonation of residues far from the active site can have on the active site structure and hence the enzyme’s activity.

The LADI-III neutron diffractometer at the Institut Laue-Langevin. © Institut Laue-Langevin / B. Lehn.
What impact can these findings have on the design of future HIV drugs?
There are various things we've suggested to improve the binding – some more straightforward than others. For example, some of the interactions between the enzyme and the drug are bridged by water molecules, and so one rather simple way to improve the binding is to add a bulkier group to the drug structure, displacing the water molecules to create direct and hence stronger hydrogen bonds between the enzyme and the drug molecule.
We have also seen that although interactions between the enzyme main-chain atoms and the inhibitor have been increased for darunavir relative to earlier protease inhibitors, in actual fact there is potential to increase and strengthen these interactions yet further still.
Analysis of all the hydrogen bond interactions showed that the two strongest hydrogen bonds are those involving the two catalytic aspartate residues.
Another approach would be to increase the strength of these two direct hydrogen bonds by the creation of what is known as a ‘low-barrier hydrogen bond’, in which the proton is equidistant between the donor and acceptor atoms.
This can be achieved by lowering the pKa (i.e. the tendency of a compound to lose a proton) of the hydroxyl group of the drug to make it similar to that of the residues it binds to, via the introduction of a strong electronegative atom such as fluorine.
Of course there's still a long way to go, but before we determined the neutron structures, it was felt that most avenues to improve the binding had been exhausted and yet the neutron structures indicate that this is not the case.
The extra information and level of detail provided by knowing the positions of the hydrogen atoms has allowed us to understand more deeply details about inhibitor binding and the enzyme’s sensitivity to pH, and with such details new avenues for improving protease inhibitors effectiveness can be explored.
X-ray crystallography has played a crucial role in structure-based drug design for over two decades. While its value to researchers in this field will continue for many years, combining the use of X-rays with neutrons increases the clarity of how drugs interact with their targets, providing the pharma industry with a powerful new tool to improve the performance of their products.
I believe that the future of drug design will feature a combination of the two crystallographic techniques that can provide patients with newer, more effective medicines to not only battle HIV infection, but for a whole host of other diseases as well.
Where can readers find more information?
More information regarding neutron crystallography studies can be found at the ILL webpages (https://www.ill.eu) or by contacting Matthew Blakeley by email (blakeleym[at]ill.fr) Further details of the neutron crystallography studies of HIV-1 protease can be found in:
[1] Long-range electrostatics-induced two-proton transfer captured by neutron crystallography in an enzyme catalytic site. Gerlits et al., (2016). Angew. Chem. Int. Ed. Engl. 55, 4924-4927.
[2] Joint X-ray/neutron crystallographic study of HIV-1 protease with clinical inhibitor amprenavir: insights for drug design. Weber et al., (2013). J. Med. Chem. 56, 5631-5635.
About Matthew Blakeley
Matthew Blakeley is a research scientist at the Institut Laue-Langevin (ILL) in Grenoble, France. After graduating from the University of Manchester in 1999 with a first class degree in Chemistry, Matthew completed his PhD in X-ray and neutron crystallography at the ILL/Manchester University in 2003, before undertaking postdoctoral research at the EMBL Grenoble Outstation.
Since 2007, Matthew has been the instrument scientist responsible for the neutron Laue diffractometer ‘LADI-III’ at the ILL. His research interests are neutron crystallography instrumentation and methods developments, structural chemistry and structural biology.