In this interview, News Medical speaks to Dr. Manasa Gudheti of Bruker Nano Surfaces about biological specimens and how to acquire super-resolution images of them.
The 2014 Nobel Prize in Chemistry led to a notable increase in publications utilizing super-resolution microscopy. How is super-resolution imaging different than traditional light microscopy?
Traditional light microscopy techniques such as confocal and wide-field are diffraction-limited in resolution, which is about 200 nm laterally (in xy) and 500 to 600 nm axially (in z). Features that are closer than the diffraction limit will appear blurred in the image.
Super-resolution techniques either surpass or circumvent the diffraction limit and thereby enable the imaging of structures in biological samples that could previously only be observed with electron microscopy (EM). In addition, live specimen imaging is possible with super-res techniques at sub-diffraction resolutions which is nearly impossible with EM.
So there are a number of different methods then to achieve super resolution. Can you briefly describe the major differences between them, and tell us what Bruker’s Vutara 352 uses?
Broadly there are three different flavors of super-resolution microscopy; Structured illumination microscopy (SIM), Stimulated emission depletion (STED) microscopy and Single Molecule Localization (SML) microscopy.
SIM can achieve a two-fold improvement in resolution using Moiré patterns and Fourier transforms. STED uses point spread function engineering to reduce the size of the diffraction limited spot achieving a five- to ten-fold improvement in resolution. SML techniques rely on localizing the positions of a series stochastically activated subset of molecules in the sample thereby achieving resolutions up to 20 nm laterally and 50 nm axially.

The Vutara 352 microscope uses SML principles coupled with biplane detection to obtain three-dimensional super-resolution images. In comparison to SIM and STED, SML techniques offer the best improvement in resolution along with quantitative/statistical analysis capabilities
What are the advantages of being able image at video rates in super resolution?
The biggest advantage would be the ability to capture dynamics in a living specimen at sub-diffraction resolution. In a fixed specimen, imaging will be faster therefore reducing recording time.
One of the major strengths of super-resolution microscopy is that it combines the three-dimensional multicolor imaging capabilities of confocal with the resolution close to those obtained by electron microscopy (EM). Even though EM slightly surpasses super-resolution techniques by obtaining near angstrom resolution, imaging live specimens with EM is nearly impossible.
Being able to image at video-rate with single-molecule techniques enables scientists to capture real-time dynamics of biological molecules in living cells. The Vutara 352 microscope makes video-rate imaging possible by using a high-speed sCMOS camera along with robust, real-time localization algorithms.
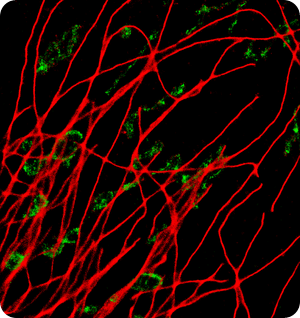
Image: BSCI cell, 800 FPS, AF647 - Alphatubulin, Cy3B - TOM20
Can the results of super-resolution imaging truly be considered quantitative, rather than qualitative?
With the right control experiments in place one can harness the quantitative aspect of single-molecule super-resolution microscopy. In the process of detecting single molecules, SML techniques are inherently counting molecules to build a map of localized molecules to render the final image. This enables statistical analysis on the tabulated co-ordinates thereby providing quantitative information in addition to the super-resolution image.
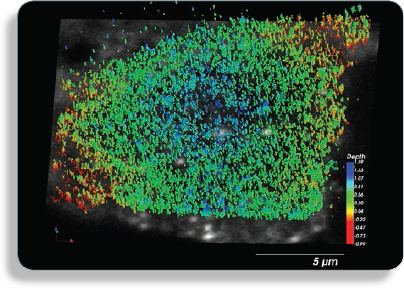
Image: Alexafluor-633 labeled GPI-protein GFP-FR, plasma membrane protein that forms distinct clusters.
Where do you think super-resolution imaging will have the biggest impact, and what is next for this exciting technology?
I think we are just beginning to scratch the surface in terms of possibilities for this technology. Recently, we added a multi-point scanning confocal scanner to the Vutara 352 microscope. This enables researchers to perform correlative imaging; i.e., overlay a super-resolution image over a contextual confocal image.
The biggest impact to come will be seen in live-cell and tissue imaging. There have been several challenges in extending SML imaging to tissues and live specimens but I think we are seeing considerable progress being made in dye engineering and sample prep methods that are making it possible.
Do you have any case studies you can tell us about?
Researchers have used the Vutara microscope to image a variety of biological specimens including cells, viruses, bacteria, tissue sections, Drosophila and C. Elegans. The Bruker website features some examples of applications: https://www.bruker.com/products/fluorescence-microscopes/vutara-super-resolution-microscopy/applications.html
There are also several publications with super-resolution data taken on the Vutara microscope.
Where can our readers learn more about super-resolution imaging?
The super-resolution field is continuously evolving and we at Bruker keep a close eye on current literature. Please head on over to our key publication page to learn more: https://www.bruker.com/en.html
About Dr. Manasa Gudheti
 Dr. Manasa Gudheti is an applications scientist at Bruker Nano Surfaces in Salt Lake City, UT, with over a decade of expertise in super-resolution microscopy.
Dr. Manasa Gudheti is an applications scientist at Bruker Nano Surfaces in Salt Lake City, UT, with over a decade of expertise in super-resolution microscopy.
Manasa earned a Bachelor’s degree in Chemical Engineering from the National Institute of Technology, Warangal, India in 1999 and a Ph.D. in Chemical Engineering from Drexel University in 2004. She then pursued a post-doctorate in Biophysics in Dr. Sam Hess’s lab at the University of Maine.
In 2006, Dr. Hess’ lab pioneered the SML microscopy technique FPALM (fluorescent photoactivation localization microscopy). In 2008, 3D FPALM was developed. This 3D super-resolution technique was subsequently commercialized at the University of Utah startup venture, Vutara. Manasa accepted the senior scientist position at Vutara in 2011 along with a visiting researcher affiliation at the University of Utah.
Bruker acquired Vutara in July, 2014. This was also the year that super-resolved fluorescence microscopy techniques garnered the Nobel Prize in Chemistry. Bruker's flagship super-resolution product, which Manasa has been instrumental in developing, is the Vutara 352 single molecule localization (SML) 3D super-resolution microscope, based on a patented biplane technology.
Vutara Publication List
- Pengpeng Li, Sean A. Merrill, Erik M. Jorgensen, Kang Shen. Two Clathrin Adaptor Protein Complexes Instruct Axon-Dendrite Polarity. (2016) Neuron, 90(3):564-580. doi: 10.1016/j.neuron.2016.04.020.
- Lukáš Alán, Tomáš Špaček, Petr Ježek. Delaunay algorithm and principal component analysis for 3D visualization of mitochondrial DNA nucleoids by Biplane FPALM/dSTORM. (2016) Eur Biophys J. DOI 10.1007/s00249-016-1114-5.
- Jonathan M. Hartley, Rui Zhang, Manasa Gudheti, Jiyuan Yang, Jindřich Kopeček. Tracking and quantifying polymer therapeutic distribution on a cellular level using 3D dSTORM. (2016) J Control Release. S0168-3659(16)30058-X. doi: 10.1016/j.jconrel.2016.02.005.
- Tomáš Olejár, David Pajuelo‑Reguer, Lukáš Alán, Andrea Dlasková, Petr Ježek. (2015). Coupled aggregation of mitochondrial single-strand DNA-binding protein tagged with Eos fluorescent protein visualizes synchronized activity of mitochondrial nucleoids. (2015) Molecular Medicine Reports, 12, 5185-5190. http://dx.doi.org/10.3892/mmr.2015.4085
- Hisashi Akiyama, Nora-Guadalupe Pina Ramirez, Manasa V. Gudheti, Suryaram Gummuluru. CD169-mediated trafficking of HIV to plasma membrane invaginations in dendritic cells attenuates efficacy of anti-gp120 broadly neutralizing antibodies. (2015) PLoS Pathog. 11(3):e1004751.
- Gabriella Kiss, Jens M. Holl, Grant M. Williams, Eric Alonas, Daryll Vanover, Aaron W. Lifland, Manasa Gudheti, Ricardo C. Guerrero-Ferreira, Vinod Nair, Hong Yi, Barney S. Graham, Philip J. Santangelo, and Elizabeth R. Wright. Structural Analysis of Respiratory Syncytial Virus Reveals the Position of M2-1 Between the Matrix Protein and the Ribonucleoprotein Complex. (2014) J. Virol. 88(13): 7602-7617
- Rui Zhang, Jiyuan Yang, Monika Sima, Yan Zhou, Jindrich Kopeček. Sequential combination therapy of ovarian cancer with degradable N-(2-hydroxypropyl)methacrylamide copolymer paclitaxel and gemcitabine conjugates. (2014) Proc Natl Acad Sci U S A. 111(33):12181-12186.
- Eric Alonas, Aaron W. Lifland, Manasa Gudheti, Daryll Vanover, Jeenah Jung, Chiara Zurla, Jonathan Kirschman, Vincent F. Fiore, Alison Douglas, Thomas H. Barker, Hong Yi, Elizabeth R. Wright, James E. Crowe , Jr., and Philip J. Santangelo. Combining Single RNA Sensitive Probes with Subdiffraction-Limited and Live-Cell Imaging Enables the Characterization of Virus Dynamics in Cells. (2014) ACS Nano. 8 (1): 302–315.
- Justin G. Lichter, Eric Carruth, Chelsea Mitchell, Andreas S. Barth, Takeshi Aiba, David A. Kass, Gordon F. Tomaselli, John H. Bridge, and Frank B. Sachse. Remodeling of the sarcomeric cytoskeleton in cardiac ventricular myocytes during heart failure and after cardiac resynchronization therapy. (2014) J Mol Cell Cardiol. 72C:186-195.
- Jeffery Hodges, Xiaolin Tang, Michael B. Landesman, John B. Ruedas, Anil Ghimire, Manasa V. Gudheti, Jacques Perrault, Erik M. Jorgensen, Jordan M. Gerton, Saveez Saffarian. Asymmetric packaging of polymerases within vesicular stomatitis virus. (2013) Biochem. Biophys. Res. Commun. 440(2): 271-276.
- Nicolas Olivier, Debora Keller, Pierre Gönczy, and Suliana Manley. Resolution Doubling in 3D-STORM Imaging through Improved Buffers. (2013) Plos One. 8(7): e69004.
- Brigitte Ritter, Sebastian Murphy, Hatem Dokainish, Martine Girard, Manasa V. Gudheti, Guennadi Kozlov, Marilene Halin, Jacynthe Philie, Erik M. Jorgensen, Kalle Gehring, and Peter S. McPherson. NECAP 1 Regulates AP-2 Interactions to Control Vesicle Size, Number, and Cargo During Clathrin-Mediated Endocytosis. (2013) PLoS Biol. 11(10):e1001670.
About Bruker Nano Surfaces and Metrology

Bruker’s suite of fluorescence microscopy systems provides a full range of solutions for life science researchers. Their multiphoton imaging systems provide the imaging depth, speed and resolution required for intravital imaging applications, and their confocal systems enable cell biologists to study function and structure using live-cell imaging at speeds and durations previously not possible. Bruker’s super-resolution microscopes are setting new standards with quantitative single molecule localization that allows for the direct investigation of the molecular positions and distribution of proteins within the cellular environment. And their Luxendo light-sheet microscopes, are revolutionizing long-term studies in developmental biology and investigation of dynamic processes in cell culture and small animal models.