Sponsored Content by TecanSep 27 2017
Interview conducted with Dr Bob St. Onge
Can you provide an overview of your work/your research at SGTC?
The SGTC is broadly interested in developing new technologies for biology and medical research that will ultimately improve and reduce the cost of healthcare.
My research uses budding yeast as a tool for synthetic biology, and also in chemical and functional genomic applications. We are most interested in developing methods that address difficult problems that aren’t easily addressed by existing technologies.
© toeytoey / Shutterstock.com
Can you share more about the research/experiments/assays themselves? Is there a downstream assay this feeds into, for example cDNA extraction, ID via oligo or NGS?
The assays we run typically involve measurement of thousands of yeast strains in parallel, each with a different genetic make-up. Most commonly we’re measuring growth rate or fitness of these strains during competitive growth in a single culture.
We use unique DNA barcodes to identify each strain, and following growth under a condition of interest, these barcodes are extracted from yeast and then quantified.
In the early days of this approach, we were using DNA microarrays to measure individual barcode abundance but nowadays we use Next-generation sequencing.
What impact will your research provide and what will the implications be for new studies down the road?
One impact is the generation of quantitative functional information for the genome. We can learn a lot about what a gene does by identifying conditions under which that gene is essential for viability of the organism.
In the past, these experiments involved gene knockouts, that is, strains where an entire gene was deleted. More recently though, we’re using gene-editing technologies such as CRISPR/Cas9, to generate smaller, precise genetic changes, for example, those that result in a single amino acid change, and measure the effect of those.
These types of genetic changes are much more common in nature, and understanding these genotype-phenotype links in yeast will eventually lead to a better understanding of these links in people.
We also use these assays to understand the mechanism of drugs and other bioactive small molecules. Mutations affecting the abundance of a drug’s protein target will often alter the cell’s response to that drug.
So, by systematically quantifying the fitness of thousands of strains in the presence of a drug, we can identify likely drug targets in an unbiased way. In the past, these experiments used commercial chemical libraries, but more recently we’ve been able to engineer yeast to produce our own compounds - by expressing natural product pathways from other fungi.
Yeast have certain attributes that make them a great host for assembling these pathways from synthetic DNA. Deploying our genomic assays to understand the activities of any compounds produced by these pathways is then straightforward.
Can you describe your automated process of the workflow itself?
Essentially, our process involves producing large collections of strains, growing these strains competitively under different conditions, and then quantifying the fitness of each strain in each condition. The competitive growth phase is a key part of the process.
Yeast grow very fast, doubling every 90 minutes or so, and collecting multiple samples from cultures that may be growing at different rates is virtually impossible without an automated system. These samples need to be stored appropriately until they can be processed further.
Can you provide some details on the automated system you use, both hardware and software?
The system we’re using was modelled after a system designed at the SGTC by my colleague Dr. Michael Proctor. The hardware consists of a Tecan Freedom EVO liquid handler, 4 Tecan Infinite plate readers, and 4 plate coolers which are controlled by custom software.
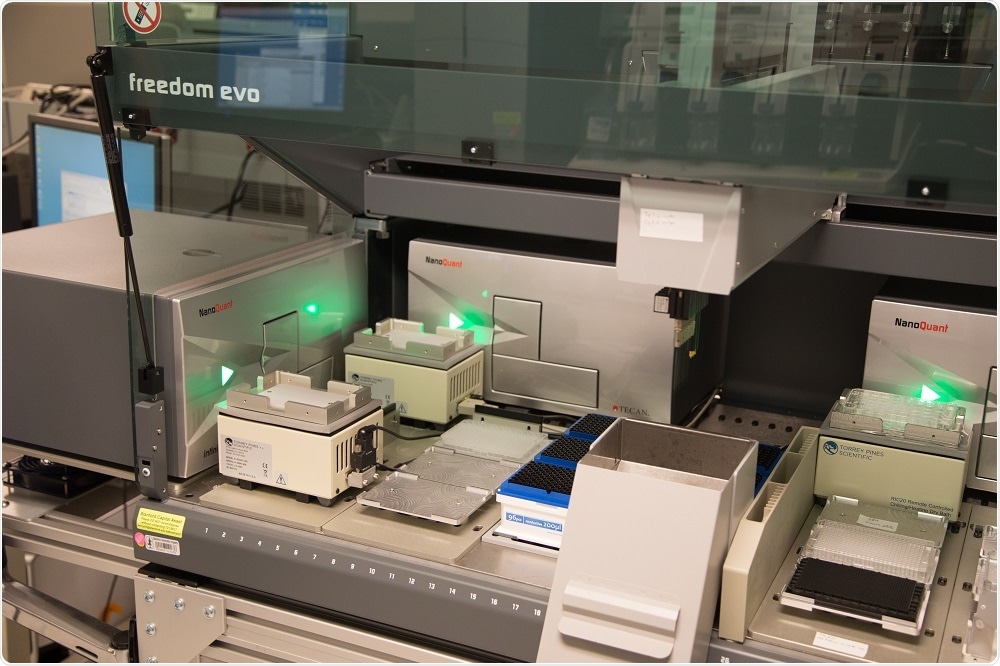
Basically, yeast are grown in microtiter plates in the Infinite readers with shaking and at a constant temperature, and growth is continuously monitored by taking optical density measurements every 15 minutes.
The system will automatically perform pipetting tasks when a ‘trigger’ OD is reached; for example, saving an aliquot of the culture to a plate cooler, adding a reagent to the culture from the deck, or diluting the culture to maintain log-phase growth over multiple generations. The user defines these tasks in the software before starting the experiment.
What are the benefits afforded by the automated system and what barriers has it helped you overcome?
There are several benefits. For one, conducting the experiments in microtiter plates, as opposed to flasks, allows us to process many more samples and minimize reagent use. In addition, collecting samples at a fixed point in growth of many different cultures can only realistically be done with an automated system like this.
This really enhances the reproducibility of the assay. Finally, the OD measurements collected provide a detailed view of growth kinetics of our pooled culture, which is often useful in interpreting the results of the DNA barcode quantification.
How satisfied are you with the performance of the system? Why Tecan EVO?
We’ve been very satisfied mainly because the system reliably executes the protocols we use most commonly. Our previous work identified some key design features, both hardware and software related, that we were able to adopt in the current system with help from Tecan. I think that helped a lot.
Are there other uses/studies the system has been adopted for? What are the plans for the future?
We’re generating many new strain collections and plan to use the system to profile these under different growth conditions. We’re also generating new chemical compounds that will hopefully exhibit interesting new biological activities. The system will be used here as well.
Where can readers find more information?
We’ve published a few papers recently where the system was used. (PMIDs: 26956608, 28193641, 28541284)
http://med.stanford.edu/sgtc/publications.html
About Dr. Bob St. Onge
Bob St.Onge is a Senior Research Scientist at the Stanford Genome Technology Center who develops novel synthetic biology and genomic methods in the budding yeast S. cerevisiae.
These methods are used to better understand genotype-phenotype relationships, define mechanism of action of bioactive compounds, and optimize heterologous expression of natural product gene clusters in yeast.