Researchers have come up with a new way to image cell populations and their genetic contents. Their study, appearing June 20 in the journal Cell, describes how a technique called DNA microscopy helps illuminate the spatial organization of genetic material within cells and tissues without specialized, expensive optical equipment. Using only the sample itself plus reagents delivered with pipettes, DNA microscopy prompts a specimen to provide spatial information about itself as part of a chemical reaction--the products of which can be read out by DNA sequencing.
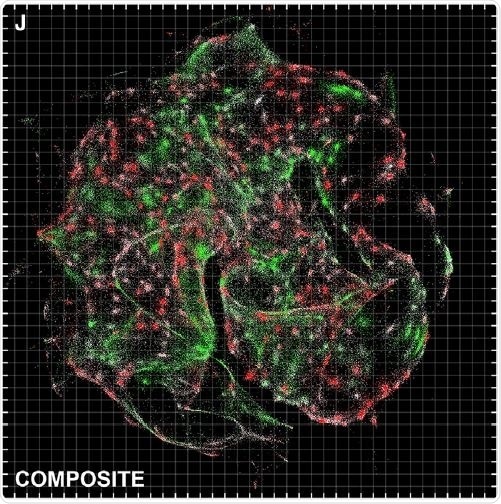
This image shows a visualization of the data provided by DNA microscopy, which has a resolution comparable to optical imaging. Credit: Weinstein et al.
If light microscopy can be compared to taking photographs of a city from an orbiting satellite, DNA microscopy is like touring that city at street level, says co-author Joshua Weinstein, a postdoctoral associate at the Broad Institute of Harvard and MIT.
Weinstein, computational and systems biologist Aviv Regev, and neuroscientist and molecular biologist Feng Zhang (@zhangf) have used DNA microscopy to image human cancer cell lines. Their goal is to accurately image long stretches of the highly variable gene sequences found in cancer mutations, immune receptors, immunoglobulin genes, and more.
Understanding how cells interact with one another is critical for advancing biological research and clinical treatments. Despite progress in profiling cells' molecular constituents, spatially mapping these constituents is still machine intensive, relying on either light microscopy or slicing and dissecting.
To understand how DNA microscopy works, imagine constructing a map of cities in the United States based on radio signals between radio towers. Even if each city's radio tower pings only its nearest neighbors, algorithms can compile this incomplete, imprecise data into an accurate map.
In DNA microscopy, a chemical reaction tags short segments of DNA called unique molecular identifiers (UMIs). The UMIs are the radio towers, and the radio signals are clouds of copies of UMIs following the physics of diffusion.
Thanks to the UMIs, the sample being studied is now dotted with chemically discrete points. Tracking the collisions between clouds of UMIs copies--with each collision written into DNA sequence products as a chemical reaction--allows researchers to narrow the uncertainty of original UMI positions. The resulting image is a two- or three-dimensional genetically detailed plot of molecular positions in physical space.
The plot represents hundreds of thousands of dimensions dictated by the number of molecules with which the tagged molecules can plausibly communicate.
A chemical reaction within the specimen encodes information into DNA from which an algorithm can decode the relative positions of molecules without needing to know in advance cell identity or the nature of genetic variation,"
Co-author Joshua Weinstein, postdoctoral associate at the Broad Institute of Harvard and MIT
DNA microscopy's weakness is resolving empty spaces, such as large gaps between two cells plated on a dish. If this can be addressed, the researchers hope to more fully explore miniscule spatial structures in the biological world, revealing layers of information that could be hidden by the limits of optical- and electron-based imaging.
"We believe that the most exciting applications of this technology are in areas of biology in which mutations, RNA editing, and other forms of nucleotide-level variation work hand in hand within the organism to either produce physiological outcomes or cause disease," Weinstein says. Examples include understanding how the immune system develops, how the nervous system is wired, and how genetic mutations are present in tumors and affect their interactions with other cells, including immune cells.
Source:
Journal reference:
Weinstein, J.A. et al. (2019) DNA Microscopy: Optics-free Spatio-genetic Imaging by a Stand-Alone Chemical Reaction. Cell. doi.org/10.1016/j.cell.2019.05.019