Geneticists have long awaited the ability to simply knock out or add a single gene here and there, as desired, with accuracy and efficiency, and then study the exact consequences caused by the loss or addition of that genetic information. In answer, the realm of genetics has been revolutionized by the emergence of the powerful gene-editing tool called CRISPR-Cas9, which allows scientists to insert or remove single genes with ease and precision across a wide range of cells.
3d illustration of CRISPR-Cas9 genome editing system - Illustration Credit: Meletios Verras / Shutterstock
In an exciting new study, CRISPR-Cas9 use is followed by laser microdissection and single cell genotyping. This protocol makes it possible to find out exactly what phenotypic changes the CRISPR-induced mutations caused in the cell studied, and then to confirm the exact nature of the underlying DNA change that produced that phenotype. This will make CRISPR still more of a dream tool for gene study, especially in the nervous system.
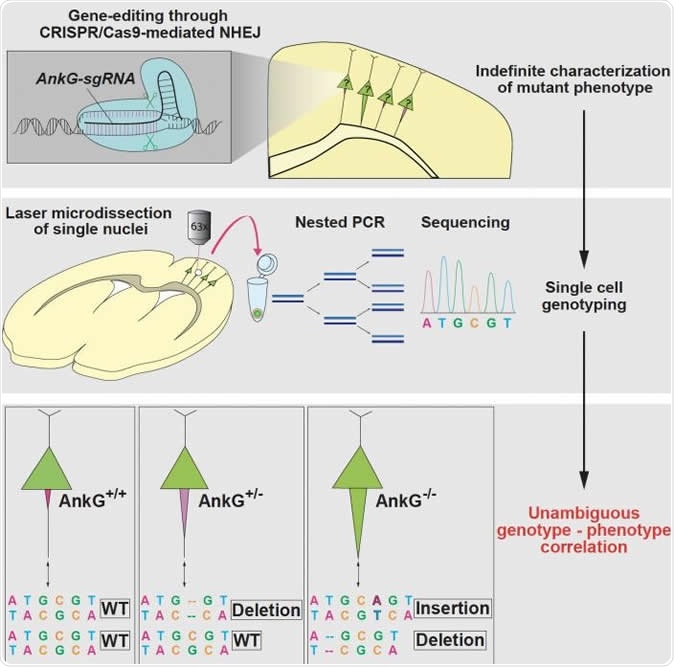
CRISPR/Cas9-based mutagenesis through NHEJ causes a variety of genotypes in individual cells, which make it difficult to determine the causality between genotypes and phenotypes. Steinecke et al. report a strategy for single-cell genotyping in CRISPR/Cas9-transfected neurons that were phenotypically characterized in vivo. Image Credit: Max Planck Florida Institute for Neuroscience
CRISPR-Cas9 does its job by cutting the DNA strand in the cell at the target site, following which the cell repairs the strand using a process called nonhomologous end joining (NHEJ). The problem is that NHEJ is not a very precise process, and can itself introduce new material into the DNA strand, or leave some material out, or substitute one small piece of DNA for another. These are all different types of additional mutations that the researchers do not want in that cell.
Another challenge is that CRISPR doesn’t always achieve a complete knockout of the gene targeted in both copies of the gene. If even one copy is still functioning, the phenotype-genotype match becomes difficult. In short, unknown mutations make it difficult to tell if the phenotype observed is due to the known mutation they introduced via CRISPR or the inadvertent changes caused by NHEJ or CRISPR inefficiency itself. So far there was no way to determine the genotype of single cells that had undergone CRISPR-Cas9 transfection. Immunohistochemistry is widely used to confirm that the target gene has been deleted, but it depends on whether the right antibodies are available for the specific proteins encoded by the targeted genes. Even then, the antibodies may not distinguish defective nonfunctional proteins from the intact kind. This makes it difficult to verify whether knockout of the gene truly occurred or not.
The chief importance of the new experimental pipeline is to study the effect of genes which do not produce overt effects or which are not easily observed. This is often the case in the brain neurons. First author Andre Steinecke says, “Some, especially genes in the brain, don't have strikingly obvious effects or are very difficult to visualize. Our goal was to create a widely applicable strategy, capable of reliably determining the exact genetic cause and correlate it to observed phenotype.”
The researchers used a gene that produces a structural protein called Ankyrin-G (AnkG) within specific brain neurons called pyramidal neurons (PN). This protein is found within the axon initial segment (AIS) of the nerve cell, and if it is not produced, the AIS shows thickening that is easily recognizable at microscopic scale. This allows neurons which do and do not produce AnkG to be easily differentiated, so that the difference in their genotypes can be clearly specified.
In the study, it was found that most CRISPR-treated neurons lost AnkG activity and showed the characteristic AIS thickening. Some of them, however, still had normal AnkG levels and AIS thickness, showing how different effects are produced by CRISPR on different cells.
With these phenotypes in mind, the brain tissue slices were now re-sectioned to achieve much thinner slices which could be subjected to laser microdissection. This allows individual nerve cells to be carefully teased out from the preparation, their phenotypes already known. These cells were subjected to DNA extraction and sequencing at single-cell level was carried out. The results perfectly matched the observed phenotype in every case. This means that each cell’s phenotype could be linked to the genotype, to find out which mutation causes what phenotypic change, with the utmost reliability and reproducibility. The presence of the mutation in one or both copies of the gene is also confirmed. In this case, the absence of AnkG and the presence of AIS thickening were linked to a loss-of-function mutation of the AnkG gene, while normal levels of AnkG were correlated with mutations in only one copy of the gene (monoallelic mutation) or no mutations.
The process was then repeated using two more genes, MeCP2 and Satb2, to confirm the protocol could indeed link phenotype to genotype with efficiency and accuracy. This protocol has added to the impressive power of CRISPR-Cas9 technology to study genes in relation to their observed effects. This is particularly useful when the cell under study doesn’t produce obvious external changes, but must be compared with other cells of the same type which have not undergone mutation.
Hiroki Taniguchi sums it up: “Our strategy will promote in vivo functional analysis of cortical genes by CRISPR/Cas-based technologies and contribute to understanding the molecular bases for function and dysfunction of cortical circuits. [It will] also be useful to other organs and tissues as well as organoids and/or explants."
The study was carried out at the Max Planck Florida Institute for Neuroscience, and was published in the journal Cell Reports on July 9, 2019.
Journal reference:
André Steinecke, Nobuhiro Kurabayashi, Yasufumi Hayano, Yugo Ishino, Hiroki Taniguchi, 'In Vivo Single-Cell Genotyping of Mouse Cortical Neurons Transfected with CRISPR/Cas9', Cell Reports, DOI: https://doi.org/10.1016/j.celrep.2019.06.038