Antibiotic resistance is a rapidly progressing phenomenon, causing huge headaches for public health planners. However, a new study demonstrates a novel strategy to kill bacteria using natural bacteria-infecting viruses called bacteriophages, or phages for short, which kill off their hosts. The bottleneck for researchers has been rapidly identifying and modifying known phages to weaponize against infectious bacteria. The new approach deals with this by using a single common framework upon which modifications can be made.
 Design_Cells | Shutterstock
Design_Cells | Shutterstock
Phages and their actions
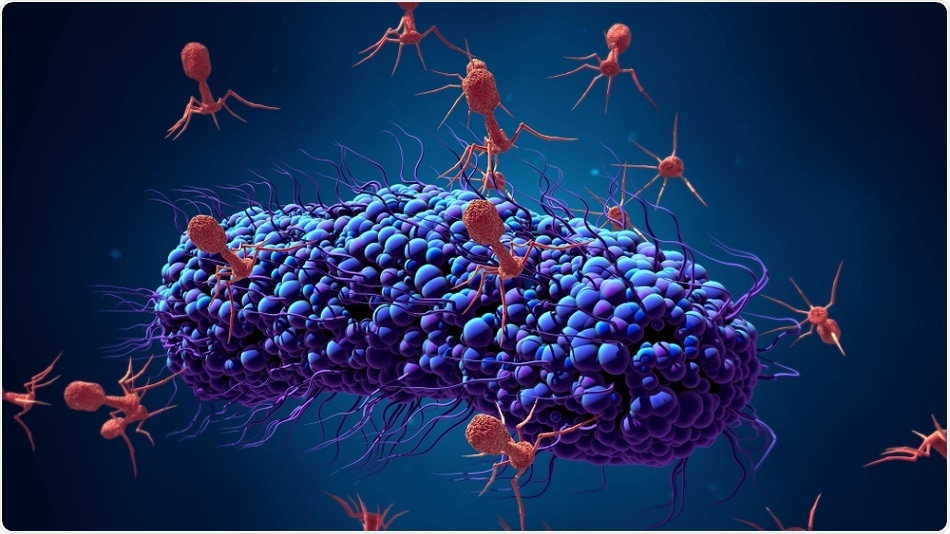
Phages specifically identify target bacteria, attach to them by their tail fibers, which results in rapid lysis or destruction of the bacterial cell wall. This property is invaluable for an antibacterial attack using antibiotic-independent mechanisms. This could mean they can destroy bacterial strains that are resistant to multiple drugs. Moreover, they are focused on the bacterium itself, avoiding off-target or unintentional damage to the body. They leave normal healthy bacteria alone, cause no significant toxicity due to their biological nature, and allow the development process to be slimmer and cheaper. They replicate themselves but disappear once the task is completed.
Phages have been around since 1915, and their use as antibacterials is also very old. Despite good early results, the lack of good trial design, and the variable results that were obtained, as well as the occurrence of the antibiotic revolution, combined to push phages out of the limelight. However, drug resistance to several commonly used antibiotics has stimulated new interest in this highly specific and effective antibacterial therapy.
Phage engineering is necessary to enhance the diversity of phages available for attack by inserting gene modifications on a common scaffold tail fiber structure. It also helps overcome bacterial mechanisms that cause phage resistance, and disrupt bacterial biofilms that weaken the phage’s capability to attach to target bacteria as well as so-called antipathogenic mechanisms aimed at reducing bacterial virulence or survival potential.
Catching the phage’s tail
The first such phage was put into action in 2015. This was a T7 phage that is directed against E. coli in nature. The scientists found that they could insert specific genes into the T7 phage that encode other types of tail fibers –the protein that helps phages to bind specifically to cell surface receptors on the target host cell. This would allow them to modify the phage to make it selective against any chosen bacteria.
However, the engineering process was neither efficient nor speedy. Thus they kept on working to refine a method that would allow them to create tailor-made phages to fight any desired bacterium. One approach was successful and forms the basis of the current phage-based attack strategy.
The current study presents a strategy to quickly modify the genetic makeup of phage to stimulate the killing of various distinct strains of the intestinal bacterium Escherichia coli (E. coli). The MIT researchers behind this study produced mutations in the tail fiber protein by which the virus locks to the bacterial cell. These mutations can be tailored to broaden the host range against which the phage can act, and simultaneously reduce the ability of the bacterium to trigger protective mechanisms against phage infection.
The tail fiber approach is based on the ability of this technique to both synthesize and test a large number of tail fiber types. The tail fiber structure is already known from prior research, and consists of proteins in an arrangement called a beta-sheet. The beta-sheets within the protein are linked by loops.
The scientists launched a systematic program of changing amino acids within the protein segment that forms the loops while leaving the beta-sheets intact. This left the protein structure largely unchanged while still allowing them to tinker with well-defined regions of the tail fiber that determine how the phage interacts with the bacterium.
Finally, they came up with approximately 10 million variants of tail fibers, which were screened using several different E. coli strains that showed resistance to infection with the original phage. For instance, one strain had mutant lipopolysaccharide (LPS) receptor genes, which led to either missing or short receptors. This made them resistant to natural phages, but susceptible to some of the engineered phages. The proof came from several modified phages that were lethal to cultured E. coli. Among these, one phage even successfully eradicated two strains in infected mouse skin, which resisted natural phage killing.
Phages represent a very different way of killing bacteria than antibiotics, which is complementary to antibiotics, rather than trying to replace them.”
Researcher Timothy Lu
The team is looking forward to using the same strategy to overcome other mechanisms of phage resistance seen with E. coli. This will give rise to new phage strains against other harmful organisms as well. Moreover, phages are effective tools to combat health problems caused by certain specific gut bacteria. And since the number of phages and bacterial targets is large, “this is just the beginning,” as Yehl says.
The future for phage antibacterials
A few phages have already been approved by the US Food and Drug Administration to eradicate pathogenic bacteria from food, but their use in treating infectious disease is limited due to the difficulty of identifying appropriate phages selective for the targeted bacteria. This is why the current research is focused on developing a viral scaffold or basic framework that can be easily modified as required to kill a different bacterial strain or to overcome other types of bacterial phage resistance. The chief advantage, in Lu’s words, is that phages provide a basic weapon that can be customized for “killing and knocking down bacteria levels inside a complex ecosystem in a targeted way.”
The disadvantages that need to be handled in phage-based antibacterial action include their large size and immunogenicity, which causes rapid recognition and loss of efficacy. Moreover, gene transfer between bacterial hosts may allow the emergence of new phage-resistant bacteria, and even worse, antibiotic resistance or increased ability to colonize and infect human hosts.
Journal reference:
Phage-derived antibacterials: harnessing the simplicity, plasticity, and diversity of phages. Bio Kim, Eun Sook Kim, Yeon-Ji Yoo, Hee-Won Bae, In-Young Chung, and You-Hee Cho. Viruses. 2019 Mar; 11(3): 268. Published online 2019 Mar 18. doi: 10.3390/v11030268. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6466130/